| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:40:19 UTC |
|---|
| Updated at | 2020-11-18 16:38:47 UTC |
|---|
| CannabisDB ID | CDB004822 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | NADP |
|---|
| Description | NADP, also known as TPN or codehydrase II, belongs to the class of organic compounds known as catechols. Catechols are compounds containing a 1,2-benzenediol moiety. NADP is a strong basic compound (based on its pKa). In humans, NADP is involved in the metabolic disorder called familial hypercholanemia (fhca). Enzymes that use NADP(H) as a coenzyme Adrenodoxin reductase: This enzyme is present ubiquitously in most organisms. NADPH is produced from NADP+. NADPH is also used for anabolic pathways, such as cholesterol synthesis and fatty acid chain elongation. NADP is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Codehydrase II | ChEBI | | Codehydrogenase II | ChEBI | | Coenzyme II | ChEBI | | NAD phosphate | ChEBI | | NADP nicotinamide-adenine-dinucleotide phosphATE | ChEBI | | TPN | ChEBI | | Triphosphopyridine nucleotide | ChEBI | | NAD phosphoric acid | Generator | | NADP nicotinamide-adenine-dinucleotide phosphoric acid | Generator | | NADPH | MeSH | | Nicotinamide-adenine dinucleotide phosphate | MeSH | | Nucleotide, triphosphopyridine | MeSH | | Dinucleotide phosphate, nicotinamide-adenine | MeSH | | Nicotinamide adenine dinucleotide phosphate | MeSH | | Phosphate, nicotinamide-adenine dinucleotide | MeSH | | NADP | ChEBI, MeSH | | Adenine-nicotinamide dinucleotide phosphate | HMDB | | NADP+ | HMDB | | beta-NADP | HMDB | | beta-Nicotinamide adenine dinucleotide phosphate | HMDB | | beta-TPN | HMDB | | β-NADP | HMDB | | β-Nicotinamide adenine dinucleotide phosphate | HMDB | | β-TPN | HMDB |
|
|---|
| Chemical Formula | C21H28N7O17P3 |
|---|
| Average Molecular Weight | 743.41 |
|---|
| Monoisotopic Molecular Weight | 743.0755 |
|---|
| IUPAC Name | 1-[(2R,3R,4S,5R)-5-{[({[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl phosphono)oxy]methyl}-3,4-dihydroxyoxolan-2-yl]-3-carbamoyl-1lambda5-pyridin-1-ylium |
|---|
| Traditional Name | 1-[(2R,3R,4S,5R)-5-[({[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-hydroxy-4-(phosphonooxy)oxolan-2-yl]methoxy(hydroxy)phosphoryl phosphono}oxy)methyl]-3,4-dihydroxyoxolan-2-yl]-3-carbamoyl-1lambda5-pyridin-1-ylium |
|---|
| CAS Registry Number | 53-59-8 |
|---|
| SMILES | NC(=O)C1=C[N+](=CC=C1)[C@@H]1O[C@H](COP([O-])(=O)OP(O)(=O)OC[C@H]2O[C@H]([C@H](OP(O)(O)=O)[C@@H]2O)N2C=NC3=C2N=CN=C3N)[C@@H](O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C21H28N7O17P3/c22-17-12-19(25-7-24-17)28(8-26-12)21-16(44-46(33,34)35)14(30)11(43-21)6-41-48(38,39)45-47(36,37)40-5-10-13(29)15(31)20(42-10)27-3-1-2-9(4-27)18(23)32/h1-4,7-8,10-11,13-16,20-21,29-31H,5-6H2,(H7-,22,23,24,25,32,33,34,35,36,37,38,39)/t10-,11-,13-,14-,15-,16-,20-,21-/m1/s1 |
|---|
| InChI Key | XJLXINKUBYWONI-NNYOXOHSSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as (5'->5')-dinucleotides. These are dinucleotides where the two bases are connected via a (5'->5')-phosphodiester linkage. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | (5'->5')-dinucleotides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | (5'->5')-dinucleotides |
|---|
| Alternative Parents | |
|---|
| Substituents | - (5'->5')-dinucleotide
- Purine nucleotide sugar
- Purine ribonucleoside 2',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Nicotinamide-nucleotide
- Pyridine nucleotide
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Imidazopyrimidine
- Nicotinamide
- Purine
- Aminopyrimidine
- Monoalkyl phosphate
- Monosaccharide
- N-substituted imidazole
- Organic phosphoric acid derivative
- Pyrimidine
- Alkyl phosphate
- Phosphoric acid ester
- Pyridinium
- Pyridine
- Imidolactam
- Imidazole
- Tetrahydrofuran
- Heteroaromatic compound
- Azole
- Secondary alcohol
- Carboximidic acid derivative
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Carboximidic acid
- Primary amine
- Alcohol
- Organopnictogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Amine
- Organonitrogen compound
- Organooxygen compound
- Organic oxide
- Organic zwitterion
- Hydrocarbon derivative
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | NADP, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_1_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_1_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_1_6, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_1_7, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_6, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_7, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_8, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_9, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_10, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_11, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_12, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_13, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_14, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_15, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_16, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_17, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | NADP, TMS_2_18, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00dl-0219003700-2f52e3c5db41066a112c | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0uk9-0301009000-a3d0c464e56f6e320c80 | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00dl-0000090000-c18a7719161c63a71938 | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0udi-0000009000-1dde5b221786fe375304 | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00dl-0209000700-02057593d6470f6a2075 | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0uk9-0301109000-bf4996084c09a9176489 | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0uk9-0301009000-b3e8d304dfdc4711e0ce | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0udi-0000009000-384d275a638928dd40f1 | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00dl-0000229600-bd164bae0cc54f45b961 | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-0027900000-25e0180b7638ed18e207 | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0a4i-0011953000-c328377f842fac60b55e | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-0000009000-0ec3670e706b086d382a | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00dl-0000109700-df2a731fd5f55e9e7a8e | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0a4i-0011953000-981d13d6b50f18ebc910 | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-0000009000-f9526262833104be0a2e | 2019-11-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-0000009000-c0cedd489befb2073370 | 2019-11-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0000000900-d510a9264bee14bd85ee | 2017-07-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000t-0000000900-fb29571fb864b5c4179b | 2017-07-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-006y-2910001000-5066541857e9b5d8bbbf | 2017-07-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-1000000900-8cdcef7ab2fa3bdd51e1 | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0007-6100000900-396416b5dd4d0892e43d | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-005a-9100000000-e5765145791ae2971a89 | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0900001200-ff4ca74a6b585e359275 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0901113400-26884ea59b81b6581275 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000j-2902210000-acc59733c27dab4bbf95 | 2021-09-23 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Glucose-Alanine Cycle | 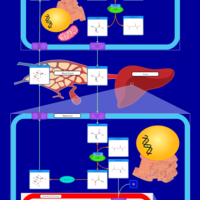 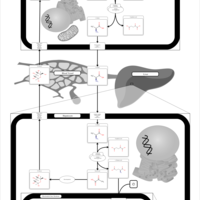 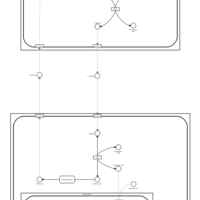 | Not Available | | Nicotinate and Nicotinamide Metabolism | 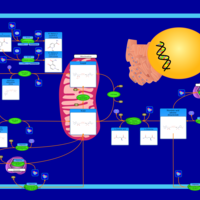 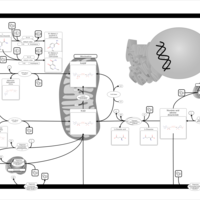 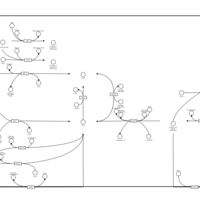 |  | | Glutathione Metabolism | 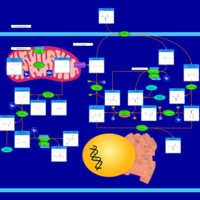 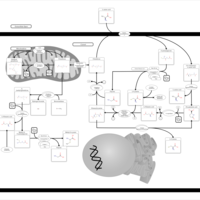 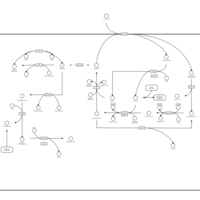 |  | | Histidine Metabolism | 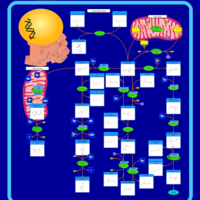 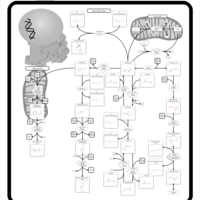 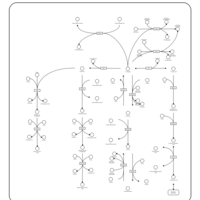 |  | | Glycerolipid Metabolism | 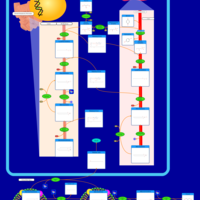 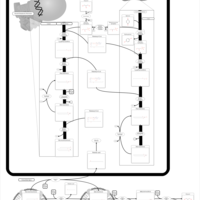 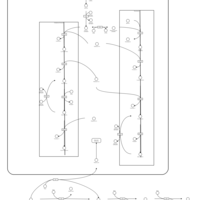 |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Fatty acid synthase | FASN | 17q25 | P49327 | details | | Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial | ALDH6A1 | 14q24.3 | Q02252 | details | | Aldo-keto reductase family 1 member C4 | AKR1C4 | 10p15.1 | P17516 | details | | Carbonyl reductase [NADPH] 1 | CBR1 | 21q22.13 | P16152 | details | | Dehydrogenase/reductase SDR family member 4 | DHRS4 | 14q11.2 | Q9BTZ2 | details | | Carbonyl reductase [NADPH] 3 | CBR3 | 21q22.2 | O75828 | details | | Aldose reductase | AKR1B1 | 7q35 | P15121 | details | | Thioredoxin reductase 1, cytoplasmic | TXNRD1 | 12q23-q24.1 | Q16881 | details | | Lathosterol oxidase | SC5DL | 11q23.3 | O75845 | details | | Dimethylaniline monooxygenase [N-oxide-forming] 5 | FMO5 | 1q21.1 | P49326 | details | | Squalene monooxygenase | SQLE | 8q24.1 | Q14534 | details | | Dimethylaniline monooxygenase [N-oxide-forming] 2 | FMO2 | 1q24.3 | Q99518 | details | | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial | ALDH4A1 | 1p36 | P30038 | details | | Thioredoxin reductase 2, mitochondrial | TXNRD2 | 22q11.21 | Q9NNW7 | details | | Dimethylaniline monooxygenase [N-oxide-forming] 4 | FMO4 | 1q24.3 | P31512 | details | | Dimethylaniline monooxygenase [N-oxide-forming] 3 | FMO3 | 1q24.3 | P31513 | details | | NADPH--cytochrome P450 reductase | POR | 7q11.2 | P16435 | details | | Dimethylaniline monooxygenase [N-oxide-forming] 1 | FMO1 | 1q24.3 | Q01740 | details | | Methylenetetrahydrofolate reductase | MTHFR | 1p36.3 | P42898 | details | | Glutathione reductase, mitochondrial | GSR | 8p21.1 | P00390 | details | | NADPH:adrenodoxin oxidoreductase, mitochondrial | FDXR | 17q24-q25 | P22570 | details | | Squalene synthase | FDFT1 | 8p23.1-p22 | P37268 | details | | Delta(14)-sterol reductase | TM7SF2 | 11q13 | O76062 | details | | Methylsterol monooxygenase 1 | MSMO1 | 4q32-q34 | Q15800 | details | | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | NSDHL | Xq28 | Q15738 | details | | Lanosterol 14-alpha demethylase | CYP51A1 | 7q21.2 | Q16850 | details | | 4-trimethylaminobutyraldehyde dehydrogenase | ALDH9A1 | 1q23.1 | P49189 | details | | Aldehyde dehydrogenase, dimeric NADP-preferring | ALDH3A1 | 17p11.2 | P30838 | details | | Alpha-aminoadipic semialdehyde dehydrogenase | ALDH7A1 | 5q31 | P49419 | details | | Aldehyde dehydrogenase family 1 member A3 | ALDH1A3 | 15q26.3 | P47895 | details | | Aldehyde dehydrogenase, mitochondrial | ALDH2 | 12q24.2 | P05091 | details | | Fatty aldehyde dehydrogenase | ALDH3A2 | 17p11.2 | P51648 | details | | Aldehyde dehydrogenase X, mitochondrial | ALDH1B1 | 9p11.1 | P30837 | details | | Pyrroline-5-carboxylate reductase 1, mitochondrial | PYCR1 | 17q25.3 | P32322 | details | | Pyrroline-5-carboxylate reductase 2 | PYCR2 | 1q42.12 | Q96C36 | details | | L-xylulose reductase | DCXR | 17q25.3 | Q7Z4W1 | details | | Glucose-6-phosphate 1-dehydrogenase | G6PD | Xq28 | P11413 | details | | Aldo-keto reductase family 1 member C3 | AKR1C3 | 10p15-p14 | P42330 | details | | GDP-mannose 4,6 dehydratase | GMDS | 6p25 | O60547 | details | | Aldo-keto reductase family 1 member C1 | AKR1C1 | 10p15-p14 | Q04828 | details | | Aldo-keto reductase family 1 member C2 | AKR1C2 | 10p15-p14 | P52895 | details | | Glyoxylate reductase/hydroxypyruvate reductase | GRHPR | 9q12 | Q9UBQ7 | details | | Methionine synthase reductase | MTRR | 5p15.31 | Q9UBK8 | details | | Sepiapterin reductase | SPR | 2p14-p12 | P35270 | details | | Estradiol 17-beta-dehydrogenase 2 | HSD17B2 | 16q24.1-q24.2 | P37059 | details | | Estradiol 17-beta-dehydrogenase 8 | HSD17B8 | 6p21.3 | Q92506 | details | | Trans-1,2-dihydrobenzene-1,2-diol dehydrogenase | DHDH | 19q13.3 | Q9UQ10 | details | | 11-cis retinol dehydrogenase | RDH5 | 12q13-q14 | Q92781 | details | | Cholesterol 7-alpha-monooxygenase | CYP7A1 | 8q11-q12 | P22680 | details | | Alcohol dehydrogenase [NADP(+)] | AKR1A1 | 1p33-p32 | P14550 | details | | Aldehyde dehydrogenase family 3 member B2 | ALDH3B2 | 11q13 | P48448 | details | | Aldehyde dehydrogenase family 3 member B1 | ALDH3B1 | 11q13 | P43353 | details | | Dihydropteridine reductase | QDPR | 4p15.31 | P09417 | details | | 7-dehydrocholesterol reductase | DHCR7 | 11q13.4 | Q9UBM7 | details | | 3-oxo-5-beta-steroid 4-dehydrogenase | AKR1D1 | 7q32-q33 | P51857 | details | | Glutamate dehydrogenase 2, mitochondrial | GLUD2 | Xq24-q25 | P49448 | details | | GMP reductase 2 | GMPR2 | 14q12 | Q9P2T1 | details | | GMP reductase 1 | GMPR | 6p23 | P36959 | details | | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | GAPDHS | 19q13.12 | O14556 | details | | Glutamate dehydrogenase 1, mitochondrial | GLUD1 | 10q23.3 | P00367 | details | | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 12p13 | P04406 | details | | 3-oxo-5-alpha-steroid 4-dehydrogenase 2 | SRD5A2 | 2p23 | P31213 | details | | 3-oxo-5-alpha-steroid 4-dehydrogenase 1 | SRD5A1 | 5p15 | P18405 | details | | Biliverdin reductase A | BLVRA | 7p13 | P53004 | details | | Flavin reductase (NADPH) | BLVRB | 19q13.1-q13.2 | P30043 | details | | Corticosteroid 11-beta-dehydrogenase isozyme 2 | HSD11B2 | 16q22 | P80365 | details | | Corticosteroid 11-beta-dehydrogenase isozyme 1 | HSD11B1 | 1q32-q41 | P28845 | details | | Dihydropyrimidine dehydrogenase [NADP(+)] | DPYD | 1p22 | Q12882 | details | | Alcohol dehydrogenase class-3 | ADH5 | 4q23 | P11766 | details | | Alcohol dehydrogenase 1B | ADH1B | 4q23 | P00325 | details | | Alcohol dehydrogenase class 4 mu/sigma chain | ADH7 | 4q23-q24 | P40394 | details | | Alcohol dehydrogenase 1A | ADH1A | 4q23 | P07327 | details | | Alcohol dehydrogenase 6 | ADH6 | 4q23 | P28332 | details | | Alcohol dehydrogenase 1C | ADH1C | 4q23 | P00326 | details | | Dihydrofolate reductase | DHFR | 5q11.2-q13.2 | P00374 | details | | Testosterone 17-beta-dehydrogenase 3 | HSD17B3 | 9q22 | P37058 | details | | Estradiol 17-beta-dehydrogenase 1 | HSD17B1 | 17q11-q21 | P14061 | details | | 3-keto-steroid reductase | HSD17B7 | 1q23 | P56937 | details | | Alpha-aminoadipic semialdehyde synthase, mitochondrial | AASS | 7q31.3 | Q9UDR5 | details | | Delta-1-pyrroline-5-carboxylate synthase | ALDH18A1 | 10q24.3 | P54886 | details | | Isocitrate dehydrogenase [NADP], mitochondrial | IDH2 | 15q26.1 | P48735 | details | | Isocitrate dehydrogenase [NADP] cytoplasmic | IDH1 | 2q33.3 | O75874 | details | | NAD-dependent protein deacetylase sirtuin-6 | SIRT6 | 19p13.3 | Q8N6T7 | details | | Ecto-ADP-ribosyltransferase 3 | ART3 | 4p15.1-p14 | Q13508 | details | | Ecto-ADP-ribosyltransferase 5 | ART5 | 11p15.4 | Q96L15 | details | | GPI-linked NAD(P)(+)--arginine ADP-ribosyltransferase 1 | ART1 | 11p15 | P52961 | details | | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | HMGCR | 5q13.3-q14 | P04035 | details | | NAD(P) transhydrogenase, mitochondrial | NNT | 5p12 | Q13423 | details | | ADP-ribosyl cyclase 2 | BST1 | 4p15 | Q10588 | details | | NAD kinase | NADK | 1p36.33 | O95544 | details | | ADP-ribosyl cyclase 1 | CD38 | 4p15 | P28907 | details | | Ecto-ADP-ribosyltransferase 4 | ART4 | 12p13-p12 | Q93070 | details | | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial | MTHFD2 | 2p13.1 | P13995 | details | | C-1-tetrahydrofolate synthase, cytoplasmic | MTHFD1 | 14q24 | P11586 | details | | GDP-L-fucose synthase | TSTA3 | 8q24.3 | Q13630 | details | | Nitric oxide synthase, inducible | NOS2 | 17q11.2-q12 | P35228 | details | | Nitric oxide synthase, brain | NOS1 | 12q24.2-q24.31 | P29475 | details | | Nitric oxide synthase, endothelial | NOS3 | 7q36 | P29474 | details | | Cytosolic 10-formyltetrahydrofolate dehydrogenase | ALDH1L1 | 3q21.3 | O75891 | details | | Trans-2-enoyl-CoA reductase, mitochondrial | MECR | 1p35.3 | Q9BV79 | details | | Peroxisomal trans-2-enoyl-CoA reductase | PECR | 2q35 | Q9BY49 | details | | Leukotriene-B(4) omega-hydroxylase 1 | CYP4F2 | 19p13.12 | P78329 | details | | Leukotriene-B(4) omega-hydroxylase 2 | CYP4F3 | 19p13.2 | Q08477 | details | | Sterol 26-hydroxylase, mitochondrial | CYP27A1 | 2q33-qter | Q02318 | details | | 6-phosphogluconate dehydrogenase, decarboxylating | PGD | 1p36.22 | P52209 | details | | 2,4-dienoyl-CoA reductase, mitochondrial | DECR1 | 8q21.3 | Q16698 | details | | Quinone oxidoreductase | CRYZ | 1p31.1 | Q08257 | details | | 25-hydroxyvitamin D-1 alpha hydroxylase, mitochondrial | CYP27B1 | 12q13.1-q13.3 | O15528 | details | | Steroid 17-alpha-hydroxylase/17,20 lyase | CYP17A1 | 10q24.3 | P05093 | details | | Peroxisomal 2,4-dienoyl-CoA reductase | DECR2 | 16p13.3 | Q9NUI1 | details | | Catalase | CAT | 11p13 | P04040 | details | | Prostaglandin reductase 1 | PTGR1 | 9q31.3 | Q14914 | details | | 25-hydroxycholesterol 7-alpha-hydroxylase | CYP7B1 | 8q21.3 | O75881 | details | | NAD(P)H dehydrogenase [quinone] 1 | NQO1 | 16q22.1 | P15559 | details | | 1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial | CYP24A1 | 20q13 | Q07973 | details | | Putative dimethylaniline monooxygenase [N-oxide-forming] 6 | FMO6P | | O60774 | details | | Delta(24)-sterol reductase | DHCR24 | 1p32.3 | Q15392 | details | | Kynurenine 3-monooxygenase | KMO | 1q42-q44 | O15229 | details | | Dehydrogenase/reductase SDR family member 9 | DHRS9 | 2q31.1 | Q9BPW9 | details | | Vitamin D 25-hydroxylase | CYP2R1 | 11p15.2 | Q6VVX0 | details | | 17-beta-hydroxysteroid dehydrogenase type 6 | HSD17B6 | 12q13 | O14756 | details | | Pyrroline-5-carboxylate reductase 3 | PYCRL | 8q24.3 | Q53H96 | details | | 3-ketodihydrosphingosine reductase | KDSR | 18q21.3 | Q06136 | details | | Aflatoxin B1 aldehyde reductase member 2 | AKR7A2 | 1p36.13 | O43488 | details | | 3 beta-hydroxysteroid dehydrogenase/Delta 5-->4-isomerase type 2 | HSD3B2 | 1p13.1 | P26439 | details | | Cholesterol 24-hydroxylase | CYP46A1 | 14q32.1 | Q9Y6A2 | details | | Retinol dehydrogenase 8 | RDH8 | 19p13.2 | Q9NYR8 | details | | Retinol dehydrogenase 11 | RDH11 | 14q24.1 | Q8TC12 | details | | 24-hydroxycholesterol 7-alpha-hydroxylase | CYP39A1 | 6p21.1-p11.2 | Q9NYL5 | details | | 7-alpha-hydroxycholest-4-en-3-one 12-alpha-hydroxylase | CYP8B1 | 3p22.1 | Q9UNU6 | details | | Retinol dehydrogenase 10 | RDH10 | 8q21.11 | Q8IZV5 | details | | Thioredoxin reductase 3 | TXNRD3 | 3q21.3 | Q86VQ6 | details | | Mitochondrial 10-formyltetrahydrofolate dehydrogenase | ALDH1L2 | 12q23.3 | Q3SY69 | details | | Fatty acyl-CoA reductase 1 | FAR1 | 11p15.2 | Q8WVX9 | details | | Fatty acyl-CoA reductase 2 | FAR2 | 12p11.22 | Q96K12 | details | | Putative uncharacterized protein DKFZp686B0215 | DKFZp686B0215 | | Q5HYI4 | details | | Monofunctional C1-tetrahydrofolate synthase, mitochondrial | MTHFD1L | 6q25.1 | Q6UB35 | details | | Kynurenine formamidase | AFMID | 17q25.3 | Q63HM1 | details | | Acyl-CoA synthetase family member 4 | AASDH | 4q12 | Q4L235 | details | | Aminoadipate-semialdehyde synthase | AASS | 7q31.3 | A4D0W4 | details | | Putative uncharacterized protein PGD | PGD | 1p36.3-p36.13 | A9Z1X1 | details | | Cytochrome P450, family 1, subfamily A, polypeptide 1 | CYP1A1 | 15q24.1 | A0N0X8 | details | | Protein-methionine sulfoxide oxidase MICAL1 | MICAL1 | 6q21 | Q8TDZ2 | details | | Protein-methionine sulfoxide oxidase MICAL2 | MICAL2 | 11p15.3 | O94851 | details | | Protein-methionine sulfoxide oxidase MICAL3 | MICAL3 | 22q11.21 | Q7RTP6 | details | | NADPH-dependent diflavin oxidoreductase 1 | NDOR1 | 9q34.3 | Q9UHB4 | details | | NADPH oxidase 1 | NOX1 | | Q9Y5S8 | details | | All-trans-retinol 13,14-reductase | RETSAT | 2p11.2 | Q6NUM9 | details | | Iodotyrosine dehalogenase 1 | IYD | 6q25.1 | Q6PHW0 | details | | Peroxisomal coenzyme A diphosphatase NUDT7 | NUDT7 | 16q23.1 | P0C024 | details | | Aldehyde dehydrogenase family 8 member A1 | ALDH8A1 | 6q23.2 | Q9H2A2 | details | | Putative L-aspartate dehydrogenase | ASPDH | 19q13.33 | A6ND91 | details | | Estradiol 17-beta-dehydrogenase 12 | HSD17B12 | 11p11.2 | Q53GQ0 | details | | Prostaglandin reductase 2 | PTGR2 | 14q24.3 | Q8N8N7 | details | | Thioredoxin domain-containing protein 2 | TXNDC2 | | Q86VQ3 | details | | Short-chain dehydrogenase/reductase 3 | DHRS3 | 1p36.1 | O75911 | details | | Polyprenol reductase | SRD5A3 | 4q12 | Q9H8P0 | details | | Very-long-chain enoyl-CoA reductase | TECR | 19p13.12 | Q9NZ01 | details | | Cytochrome P450, family 21, subfamily A, polypeptide 2 | CYP21A2 | 6p21.3 | Q08AG9 | details | | NOS1 mutant | NOS1 | 12q24.2-q24.31 | B3VK56 | details | | L-aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase | AASDHPPT | 11q22 | Q9NRN7 | details | | 14 kDa phosphohistidine phosphatase | PHPT1 | | Q9NRX4 | details | | Aldo-keto reductase family 1 member B10 | AKR1B10 | | O60218 | details | | 1,5-anhydro-D-fructose reductase | AKR1E2 | 10p15.1 | Q96JD6 | details | | Steroid 21-hydroxylase | CYP21A2 | 6p21.3 | P08686 | details | | Dihydrofolate reductase, mitochondrial | DHFRL1 | 3q11.1 | Q86XF0 | details | | NAD kinase domain-containing protein 1, mitochondrial | NADKD1 | 5p13.2 | Q4G0N4 | details |
|
|---|
| Transporters | |
| Fatty acid synthase | FASN | 17q25 | P49327 | details | | Aldo-keto reductase family 1 member C4 | AKR1C4 | 10p15.1 | P17516 | details | | Nitric oxide synthase, inducible | NOS2 | 17q11.2-q12 | P35228 | details | | Cytosolic 10-formyltetrahydrofolate dehydrogenase | ALDH1L1 | 3q21.3 | O75891 | details | | Mitochondrial 10-formyltetrahydrofolate dehydrogenase | ALDH1L2 | 12q23.3 | Q3SY69 | details | | Acyl-CoA synthetase family member 4 | AASDH | 4q12 | Q4L235 | details | | Voltage-gated potassium channel subunit beta-2 | KCNAB2 | 1p36.3 | Q13303 | details |
|
|---|
| Metal Bindings | |
| Fatty acid synthase | FASN | 17q25 | P49327 | details | | Lathosterol oxidase | SC5DL | 11q23.3 | O75845 | details | | NADPH--cytochrome P450 reductase | POR | 7q11.2 | P16435 | details | | Methylsterol monooxygenase 1 | MSMO1 | 4q32-q34 | Q15800 | details | | Lanosterol 14-alpha demethylase | CYP51A1 | 7q21.2 | Q16850 | details | | Cytochrome P450 4A11 | CYP4A11 | 1p33 | Q02928 | details | | Methionine synthase reductase | MTRR | 5p15.31 | Q9UBK8 | details | | Cholesterol 7-alpha-monooxygenase | CYP7A1 | 8q11-q12 | P22680 | details | | GMP reductase 2 | GMPR2 | 14q12 | Q9P2T1 | details | | GMP reductase 1 | GMPR | 6p23 | P36959 | details | | Biliverdin reductase A | BLVRA | 7p13 | P53004 | details | | Dihydropyrimidine dehydrogenase [NADP(+)] | DPYD | 1p22 | Q12882 | details | | Alcohol dehydrogenase class-3 | ADH5 | 4q23 | P11766 | details | | Alcohol dehydrogenase 1B | ADH1B | 4q23 | P00325 | details | | Alcohol dehydrogenase class 4 mu/sigma chain | ADH7 | 4q23-q24 | P40394 | details | | Alcohol dehydrogenase 1A | ADH1A | 4q23 | P07327 | details | | Alcohol dehydrogenase 6 | ADH6 | 4q23 | P28332 | details | | Alcohol dehydrogenase 1C | ADH1C | 4q23 | P00326 | details | | NADP-dependent malic enzyme | ME1 | 6q12 | P48163 | details | | NADP-dependent malic enzyme, mitochondrial | ME3 | 11cen-q22.3 | Q16798 | details | | NAD-dependent malic enzyme, mitochondrial | ME2 | 18q21 | P23368 | details | | Isocitrate dehydrogenase [NADP], mitochondrial | IDH2 | 15q26.1 | P48735 | details | | Isocitrate dehydrogenase [NADP] cytoplasmic | IDH1 | 2q33.3 | O75874 | details | | NAD-dependent protein deacetylase sirtuin-6 | SIRT6 | 19p13.3 | Q8N6T7 | details | | NAD kinase | NADK | 1p36.33 | O95544 | details | | Nitric oxide synthase, inducible | NOS2 | 17q11.2-q12 | P35228 | details | | Nitric oxide synthase, brain | NOS1 | 12q24.2-q24.31 | P29475 | details | | Nitric oxide synthase, endothelial | NOS3 | 7q36 | P29474 | details | | Trans-2-enoyl-CoA reductase, mitochondrial | MECR | 1p35.3 | Q9BV79 | details | | Leukotriene-B(4) omega-hydroxylase 1 | CYP4F2 | 19p13.12 | P78329 | details | | Leukotriene-B(4) omega-hydroxylase 2 | CYP4F3 | 19p13.2 | Q08477 | details | | Cytochrome P450 3A4 | CYP3A4 | 7q21.1 | P08684 | details | | Cytochrome P450 2C9 | CYP2C9 | 10q24 | P11712 | details | | Sterol 26-hydroxylase, mitochondrial | CYP27A1 | 2q33-qter | Q02318 | details | | Cytochrome P450 2C19 | CYP2C19 | 10q24 | P33261 | details | | Quinone oxidoreductase | CRYZ | 1p31.1 | Q08257 | details | | 25-hydroxyvitamin D-1 alpha hydroxylase, mitochondrial | CYP27B1 | 12q13.1-q13.3 | O15528 | details | | Steroid 17-alpha-hydroxylase/17,20 lyase | CYP17A1 | 10q24.3 | P05093 | details | | Cytochrome P450 2E1 | CYP2E1 | 10q24.3-qter | P05181 | details | | Cytochrome P450 4F11 | CYP4F11 | 19p13.1 | Q9HBI6 | details | | Cytochrome P450 3A43 | CYP3A43 | 7q21.1 | Q9HB55 | details | | Cytochrome P450 1B1 | CYP1B1 | 2p22.2 | Q16678 | details | | Cytochrome P450 2C18 | CYP2C18 | 10q24 | P33260 | details | | Cytochrome P450 2F1 | CYP2F1 | 19q13.2 | P24903 | details | | Cytochrome P450 4X1 | CYP4X1 | 1 | Q8N118 | details | | Cytochrome P450 2B6 | CYP2B6 | 19q13.2 | P20813 | details | | Cytochrome P450 3A5 | CYP3A5 | 7q21.1 | P20815 | details | | Cytochrome P450 2A13 | CYP2A13 | 19q13.2 | Q16696 | details | | Cytochrome P450 3A7 | CYP3A7 | 7q21-q22.1 | P24462 | details | | Cytochrome P450 4B1 | CYP4B1 | 1p34-p12 | P13584 | details | | Cytochrome P450 4Z1 | CYP4Z1 | 1p33 | Q86W10 | details | | Cytochrome P450 1A2 | CYP1A2 | 15q24.1 | P05177 | details | | Cytochrome P450 19A1 | CYP19A1 | 15q21.1 | P11511 | details | | Cytochrome P450 2C8 | CYP2C8 | 10q23.33 | P10632 | details | | Cytochrome P450 2S1 | CYP2S1 | 19q13.1 | Q96SQ9 | details | | Cytochrome P450 4F8 | CYP4F8 | 19p13.1 | P98187 | details | | Cytochrome P450 2J2 | CYP2J2 | 1p31.3-p31.2 | P51589 | details | | Cytochrome P450 2A7 | CYP2A7 | 19q13.2 | P20853 | details | | Cytochrome P450 2A6 | CYP2A6 | 19q13.2 | P11509 | details | | Catalase | CAT | 11p13 | P04040 | details | | Prostaglandin reductase 1 | PTGR1 | 9q31.3 | Q14914 | details | | 25-hydroxycholesterol 7-alpha-hydroxylase | CYP7B1 | 8q21.3 | O75881 | details | | Ubiquinone biosynthesis protein COQ7 homolog | COQ7 | | Q99807 | details | | 1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial | CYP24A1 | 20q13 | Q07973 | details | | Vitamin D 25-hydroxylase | CYP2R1 | 11p15.2 | Q6VVX0 | details | | Cholesterol 24-hydroxylase | CYP46A1 | 14q32.1 | Q9Y6A2 | details | | 24-hydroxycholesterol 7-alpha-hydroxylase | CYP39A1 | 6p21.1-p11.2 | Q9NYL5 | details | | 7-alpha-hydroxycholest-4-en-3-one 12-alpha-hydroxylase | CYP8B1 | 3p22.1 | Q9UNU6 | details | | Cytochrome P450 4A22 | CYP4A22 | 1p33 | Q5TCH4 | details | | Cytochrome P450, family 1, subfamily A, polypeptide 1 | CYP1A1 | 15q24.1 | A0N0X8 | details | | Protein-methionine sulfoxide oxidase MICAL1 | MICAL1 | 6q21 | Q8TDZ2 | details | | Protein-methionine sulfoxide oxidase MICAL2 | MICAL2 | 11p15.3 | O94851 | details | | Protein-methionine sulfoxide oxidase MICAL3 | MICAL3 | 22q11.21 | Q7RTP6 | details | | NADPH-dependent diflavin oxidoreductase 1 | NDOR1 | 9q34.3 | Q9UHB4 | details | | NADPH oxidase 1 | NOX1 | | Q9Y5S8 | details | | Cytochrome P450 4F22 | CYP4F22 | | Q6NT55 | details | | Peroxisomal coenzyme A diphosphatase NUDT7 | NUDT7 | 16q23.1 | P0C024 | details | | Prostaglandin reductase 2 | PTGR2 | 14q24.3 | Q8N8N7 | details | | Cytochrome P450, family 21, subfamily A, polypeptide 2 | CYP21A2 | 6p21.3 | Q08AG9 | details | | NOS1 mutant | NOS1 | 12q24.2-q24.31 | B3VK56 | details | | L-aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase | AASDHPPT | 11q22 | Q9NRN7 | details |
|
|---|
| Receptors | |
|---|
| Transcriptional Factors | |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000217 |
|---|
| DrugBank ID | DB03461 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB021908 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 5674 |
|---|
| KEGG Compound ID | C00006 |
|---|
| BioCyc ID | NADP |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Nicotinamide adenine dinucleotide phosphate |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 5885 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 44409 |
|---|
| References |
|---|
| General References | Not Available |
|---|



