| Identification |
|---|
| HMDB Protein ID
| CDBP00072 |
| Secondary Accession Numbers
| Not Available |
| Name
| Carbonyl reductase [NADPH] 1 |
| Description
| Not Available |
| Synonyms
|
- 15-hydroxyprostaglandin dehydrogenase [NADP+]
- NADPH-dependent carbonyl reductase 1
- Prostaglandin 9-ketoreductase
- Prostaglandin-E(2) 9-reductase
- 15-hydroxyprostaglandin dehydrogenase [NADP(+)]
|
| Gene Name
| CBR1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in oxidoreductase activity |
| Specific Function
| NADPH-dependent reductase with broad substrate specificity. Catalyzes the reduction of a wide variety of carbonyl compounds including quinones, prostaglandins, menadione, plus various xenobiotics. Catalyzes the reduction of the antitumor anthracyclines doxorubicin and daunorubicin to the cardiotoxic compounds doxorubicinol and daunorubicinol. Can convert prostaglandin E2 to prostaglandin F2-alpha. Can bind glutathione, which explains its higher affinity for glutathione-conjugated substrates. Catalyzes the reduction of S-nitrosoglutathione.
|
| GO Classification
|
| Biological Process |
| drug metabolic process |
| vitamin K metabolic process |
| Cellular Component |
| cytoplasm |
| Function |
| binding |
| catalytic activity |
| oxidoreductase activity |
| Molecular Function |
| oxidoreductase activity, acting on NADH or NADPH, quinone or similar compound as acceptor |
| 15-hydroxyprostaglandin dehydrogenase (NADP+) activity |
| carbonyl reductase (NADPH) activity |
| prostaglandin-E2 9-reductase activity |
| nucleotide binding |
| Process |
| oxidation reduction |
| metabolic process |
|
| Cellular Location
|
- Cytoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Metabolism of xenobiotics by cytochrome P450 | Not Available | 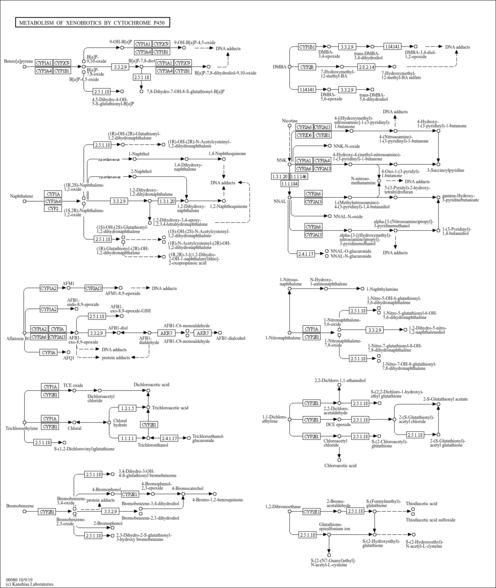 | | Chemical carcinogenesis | Not Available | 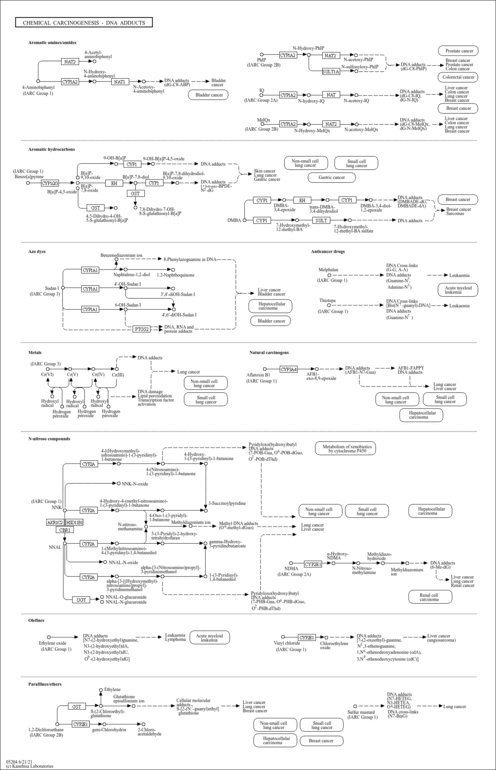 | | Arachidonic Acid Metabolism |    |  | | Leukotriene C4 Synthesis Deficiency |    | Not Available | | Piroxicam Action Pathway |    | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 21 |
| Locus
| 21q22.13 |
| SNPs
| CBR1 |
| Gene Sequence
|
>834 bp
ATGTCGTCCGGCATCCATGTAGCGCTGGTGACTGGAGGCAACAAGGGCATCGGCTTGGCC
ATCGTGCGCGACCTGTGCCGGCTGTTCTCGGGGGACGTGGTGCTCACGGCGCGGGACGTG
ACGCGGGGCCAGGCGGCCGTACAGCAGCTGCAGGCGGAGGGCCTGAGCCCGCGCTTCCAC
CAGCTGGACATCGACGATCTGCAGAGCATCCGCGCCCTGCGCGACTTCCTGCGCAAGGAG
TACGGGGGCCTGGACGTGCTGGTCAACAACGCGGGCATCGCCTTCAAGGTTGCTGATCCC
ACACCCTTTCATATTCAAGCTGAAGTGACGATGAAAACAAATTTCTTTGGTACCCGAGAT
GTGTGCACAGAATTACTCCCTCTAATAAAACCCCAAGGGAGAGTGGTGAACGTATCTAGC
ATCATGAGCGTCAGAGCCCTTAAAAGCTGCAGCCCAGAGCTGCAGCAGAAGTTCCGCAGT
GAGACCATCACTGAGGAGGAGCTGGTGGGGCTCATGAACAAGTTTGTGGAGGATACAAAG
AAGGGAGTGCACCAGAAGGAGGGCTGGCCCAGCAGCGCATACGGGGTGACGAAGATTGGC
GTCACCGTTCTGTCCAGGATCCACGCCAGGAAACTGAGTGAGCAGAGGAAAGGGGACAAG
ATCCTCCTGAATGCCTGCTGCCCAGGGTGGGTGAGAACTGACATGGCGGGACCCAAGGCC
ACCAAGAGCCCAGAAGAAGGTGCAGAGACCCCTGTGTACTTGGCCCTTTTGCCCCCAGAT
GCTGAGGGTCCCCATGGACAATTTGTTTCAGAGAAGAGAGTTGAACAGTGGTGA
|
| Protein Properties |
|---|
| Number of Residues
| 277 |
| Molecular Weight
| 30374.73 |
| Theoretical pI
| 8.319 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Carbonyl reductase [NADPH] 1
MSSGIHVALVTGGNKGIGLAIVRDLCRLFSGDVVLTARDVTRGQAAVQQLQAEGLSPRFH
QLDIDDLQSIRALRDFLRKEYGGLDVLVNNAGIAFKVADPTPFHIQAEVTMKTNFFGTRD
VCTELLPLIKPQGRVVNVSSIMSVRALKSCSPELQQKFRSETITEEELVGLMNKFVEDTK
KGVHQKEGWPSSAYGVTKIGVTVLSRIHARKLSEQRKGDKILLNACCPGWVRTDMAGPKA
TKSPEEGAETPVYLALLPPDAEGPHGQFVSEKRVEQW
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| P16152 |
| UniProtKB/Swiss-Prot Entry Name
| CBR1_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| J04056 |
| GeneCard ID
| CBR1 |
| GenAtlas ID
| CBR1 |
| HGNC ID
| HGNC:1548 |
| References |
|---|
| General References
| Not Available |


