| Identification |
|---|
| HMDB Protein ID
| CDBP00073 |
| Secondary Accession Numbers
| Not Available |
| Name
| Dehydrogenase/reductase SDR family member 4 |
| Description
| Not Available |
| Synonyms
|
- CR
- NADPH-dependent carbonyl reductase/NADP-retinol dehydrogenase
- NADPH-dependent retinol dehydrogenase/reductase
- NRDR
- PHCR
- PSCD
- Peroxisomal short-chain alcohol dehydrogenase
- SCAD-SRL
- Short-chain dehydrogenase/reductase family member 4
- humNRDR
|
| Gene Name
| DHRS4 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in oxidoreductase activity |
| Specific Function
| Reduces all-trans-retinal and 9-cis retinal. Can also catalyze the oxidation of all-trans-retinol with NADP as co-factor, but with much lower efficiency. Reduces alkyl phenyl ketones and alpha-dicarbonyl compounds with aromatic rings, such as pyrimidine-4-aldehyde, 3-benzoylpyridine, 4-benzoylpyridine, menadione and 4-hexanoylpyridine. Has no activity towards aliphatic aldehydes and ketones (By similarity).
|
| GO Classification
|
| Biological Process |
| protein tetramerization |
| steroid metabolic process |
| cellular ketone metabolic process |
| oxidation-reduction process |
| alcohol metabolic process |
| Cellular Component |
| mitochondrion |
| nucleus |
| peroxisomal membrane |
| Function |
| binding |
| catalytic activity |
| oxidoreductase activity |
| Molecular Function |
| oxidoreductase activity |
| oxidoreductase activity, acting on NADH or NADPH, quinone or similar compound as acceptor |
| alcohol dehydrogenase [NAD(P)+] activity |
| carbonyl reductase (NADPH) activity |
| 3-keto sterol reductase activity |
| nucleotide binding |
| Process |
| metabolic process |
| oxidation reduction |
|
| Cellular Location
|
- Peroxisome
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Peroxisome | Not Available | 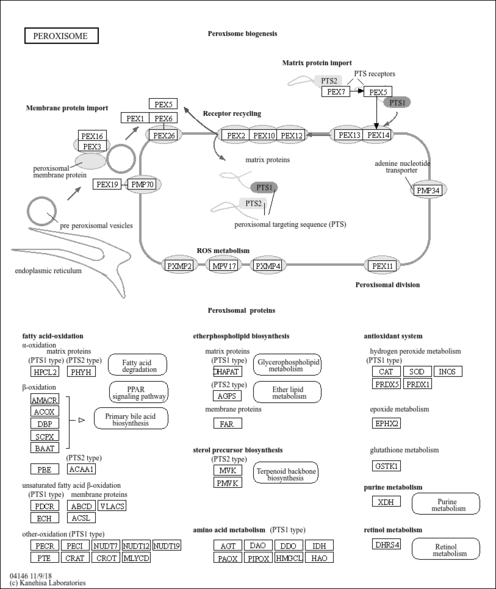 | | Retinol Metabolism |    | 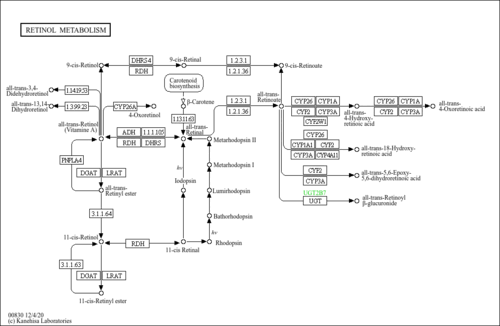 | | Vitamin A Deficiency |    | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 14 |
| Locus
| 14q11.2 |
| SNPs
| DHRS4 |
| Gene Sequence
|
>837 bp
ATGCACAAGGCGGGGCTGCTAGGCCTCTGTGCCCGGGCTTGGAATTCGGTGCGGATGGCC
AGCTCCGGGATGACCCGCCGGGACCCGCTCGCAAATAAGGTGGCCCTGGTAACGGCCTCC
ACCGACGGGATCGGCTTCGCCATCGCCCGGCGTTTGGCCCAGGACGGGGCCCATGTGGTC
GTCAGCAGCCGGAAGCAGCAGAATGTGGACCAGGCGGTGGCCACGCTGCAGGGGGAGGGG
CTGAGCGTGACGGGCACCGTGTGCCATGTGGGGAAGGCGGAGGACCGGGAGCGGCTGGTG
GCCACGGCTGTGAAGCTTCATGGAGGTATCGATATCCTAGTCTCCAATGCTGCTGTCAAC
CCTTTCTTTGGAAGCATAATGGATGTCACTGAGGAGGTGTGGGACAAGACTCTGGACATT
AATGTGAAGGCCCCAGCCCTGATGACAAAGGCAGTGGTGCCAGAAATGGAGAAACGAGGA
GGCGGCTCAGTGGTGATCGTGTCTTCCATAGCAGCCTTCAGTCCATCTCCTGGCTTCAGT
CCTTACAATGTCAGTAAAACAGCCTTGCTGGGCCTGACCAAGACCCTGGCCATAGAGCTG
GCCCCAAGGAACATTAGGGTGAACTGCCTAGCACCTGGACTTATCAAGACTAGCTTCAGC
AGGATGCTCTGGATGGACAAGGAAAAAGAGGAAAGCATGAAAGAAACCCTGCGGATAAGA
AGGTTAGGCGAGCCAGAGGATTGTGCTGGCATCGTGTCTTTCCTGTGCTCTGAAGATGCC
AGCTACATCACTGGGGAAACAGTGGTGGTGGGTGGAGGAACCCCGTCCCGCCTCTGA
|
| Protein Properties |
|---|
| Number of Residues
| 278 |
| Molecular Weight
| 29536.885 |
| Theoretical pI
| 8.555 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Dehydrogenase/reductase SDR family member 4
MHKAGLLGLCARAWNSVRMASSGMTRRDPLANKVALVTASTDGIGFAIARRLAQDGAHVV
VSSRKQQNVDQAVATLQGEGLSVTGTVCHVGKAEDRERLVATAVKLHGGIDILVSNAAVN
PFFGSIMDVTEEVWDKTLDINVKAPALMTKAVVPEMEKRGGGSVVIVSSIAAFSPSPGFS
PYNVSKTALLGLTKTLAIELAPRNIRVNCLAPGLIKTSFSRMLWMDKEKEESMKETLRIR
RLGEPEDCAGIVSFLCSEDASYITGETVVVGGGTPSRL
|
| External Links |
|---|
| GenBank ID Protein
| 32483357 |
| UniProtKB/Swiss-Prot ID
| Q9BTZ2 |
| UniProtKB/Swiss-Prot Entry Name
| DHRS4_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| NM_021004.2 |
| GeneCard ID
| DHRS4 |
| GenAtlas ID
| DHRS4 |
| HGNC ID
| HGNC:16985 |
| References |
|---|
| General References
| Not Available |

