| Identification |
|---|
| HMDB Protein ID
| CDBP01444 |
| Secondary Accession Numbers
| Not Available |
| Name
| Cytochrome P450 2A13 |
| Description
| Not Available |
| Synonyms
|
- CYPIIA13
|
| Gene Name
| CYP2A13 |
| Protein Type
| Metal Binding |
| Biological Properties |
|---|
| General Function
| Involved in monooxygenase activity |
| Specific Function
| Exhibits a coumarin 7-hydroxylase activity. Active in the metabolic activation of hexamethylphosphoramide, N,N-dimethylaniline, 2'-methoxyacetophenone, N-nitrosomethylphenylamine, and the tobacco-specific carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Possesses phenacetin O-deethylation activity.
|
| GO Classification
|
| Biological Process |
| small molecule metabolic process |
| xenobiotic metabolic process |
| Cellular Component |
| endoplasmic reticulum membrane |
| Function |
| metal ion binding |
| binding |
| catalytic activity |
| transition metal ion binding |
| electron carrier activity |
| iron ion binding |
| monooxygenase activity |
| heme binding |
| oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen |
| oxidoreductase activity |
| ion binding |
| cation binding |
| Molecular Function |
| electron carrier activity |
| aromatase activity |
| iron ion binding |
| heme binding |
| Process |
| metabolic process |
| oxidation reduction |
|
| Cellular Location
|
- Peripheral membrane protein
- Peripheral membrane protein
- Endoplasmic reticulum membrane
- Microsome membrane
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Metabolism of xenobiotics by cytochrome P450 | Not Available |  | | Drug metabolism - cytochrome P450 | Not Available |  | | Drug metabolism - other enzymes | Not Available |  | | Chemical carcinogenesis | Not Available |  | | Retinol Metabolism |    | 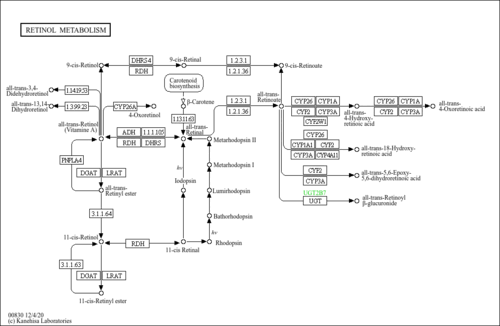 |
|
| Gene Properties |
|---|
| Chromosome Location
| 19 |
| Locus
| 19q13.2 |
| SNPs
| CYP2A13 |
| Gene Sequence
|
>1485 bp
ATGCTGGCCTCAGGGCTGCTTCTGGTGACCTTGCTGGCCTGCCTGACTGTGATGGTCTTG
ATGTCAGTCTGGCGGCAGAGGAAGAGCAGGGGGAAGCTGCCTCCGGGACCCACCCCATTG
CCCTTCATTGGAAACTACCTGCAGCTGAACACAGAGCAGATGTACAACTCCCTCATGAAG
ATCAGTGAGCGCTATGGCCCTGTGTTCACCATTCACTTGGGGCCCCGGCGGGTCGTGGTG
CTGTGCGGACATGATGCCGTCAAGGAGGCTCTGGTGGACCAGGCTGAGGAGTTCAGCGGG
CGAGGCGAGCAGGCCACCTTCGACTGGCTCTTCAAAGGCTATGGCGTGGCGTTCAGCAAC
GGGGAGCGCGCCAAGCAGCTCCGGCGCTTCTCCATCGCCACCCTAAGGGGTTTTGGCGTG
GGCAAGCGCGGCATCGAGGAACGCATCCAGGAGGAGGCGGGCTTCCTCATCGACGCCCTC
CGGGGCACGCACGGCGCCAATATCGATCCCACCTTCTTCCTGAGCCGCACAGTCTCCAAT
GTCATCAGCTCCATTGTCTTTGGGGACCGCTTTGACTATGAGGACAAAGAGTTCCTGTCA
CTGTTGCGCATGATGCTGGGAAGCTTCCAGTTCACGGCAACCTCCACGGGGCAGCTCTAT
GAGATGTTCTCTTCGGTGATGAAACACCTGCCAGGACCACAGCAACAGGCCTTTAAGGAG
CTGCAAGGGCTGGAGGACTTCATCGCCAAGAAGGTGGAGCACAACCAGCGCACGCTGGAT
CCCAATTCCCCACGGGACTTCATCGACTCCTTTCTCATCCGCATGCAGGAGGAGGAGAAG
AACCCCAACACAGAGTTCTACTTGAAGAACCTGGTGATGACCACCCTGAACCTCTTCTTT
GCGGGCACTGAGACCGTGAGCACCACCCTGCGCTACGGTTTCCTGCTGCTCATGAAGCAC
CCAGAGGTGGAGGCCAAGGTCCATGAGGAGATTGACAGAGTGATCGGCAAGAACCGGCAG
CCCAAGTTTGAGGACCGGGCCAAGATGCCCTACACAGAGGCAGTGATCCACGAGATCCAA
AGATTTGGAGACATGCTCCCCATGGGTTTGGCCCACAGGGTCAACAAGGACACCAAGTTT
CGGGATTTCTTCCTCCCTAAGGGCACTGAAGTGTTCCCTATGCTGGGCTCCGTGCTGAGA
GACCCCAGGTTCTTCTCCAACCCCCGGGACTTCAATCCCCAGCACTTCCTGGATAAGAAG
GGGCAGTTTAAGAAGAGTGATGCTTTTGTGCCCTTTTCCATCGGAAAGCGGTACTGTTTT
GGAGAAGGCCTGGCCAGAATGGAGCTCTTTCTCTTCTTCACCACCATCATGCAGAACTTT
CGCTTCAAGTCCCCTCAGTCGCCTAAGGATATCGACGTGTCCCCCAAACACGTGGGCTTT
GCCACGATCCCACGAAACTACACCATGAGCTTCCTGCCCCGCTGA
|
| Protein Properties |
|---|
| Number of Residues
| 494 |
| Molecular Weight
| 56687.095 |
| Theoretical pI
| 9.27 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Cytochrome P450 2A13
MLASGLLLVTLLACLTVMVLMSVWRQRKSRGKLPPGPTPLPFIGNYLQLNTEQMYNSLMK
ISERYGPVFTIHLGPRRVVVLCGHDAVKEALVDQAEEFSGRGEQATFDWLFKGYGVAFSN
GERAKQLRRFSIATLRGFGVGKRGIEERIQEEAGFLIDALRGTHGANIDPTFFLSRTVSN
VISSIVFGDRFDYEDKEFLSLLRMMLGSFQFTATSTGQLYEMFSSVMKHLPGPQQQAFKE
LQGLEDFIAKKVEHNQRTLDPNSPRDFIDSFLIRMQEEEKNPNTEFYLKNLVMTTLNLFF
AGTETVSTTLRYGFLLLMKHPEVEAKVHEEIDRVIGKNRQPKFEDRAKMPYTEAVIHEIQ
RFGDMLPMGLAHRVNKDTKFRDFFLPKGTEVFPMLGSVLRDPRFFSNPRDFNPQHFLDKK
GQFKKSDAFVPFSIGKRYCFGEGLARMELFLFFTTIMQNFRFKSPQSPKDIDVSPKHVGF
ATIPRNYTMSFLPR
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| Q16696 |
| UniProtKB/Swiss-Prot Entry Name
| CP2AD_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AF209774 |
| GeneCard ID
| CYP2A13 |
| GenAtlas ID
| CYP2A13 |
| HGNC ID
| HGNC:2608 |
| References |
|---|
| General References
| Not Available |




