| Identification |
|---|
| HMDB Protein ID
| CDBP00659 |
| Secondary Accession Numbers
| Not Available |
| Name
| Glyceraldehyde-3-phosphate dehydrogenase |
| Description
| Not Available |
| Synonyms
|
- GAPDH
- Peptidyl-cysteine S-nitrosylase GAPDH
|
| Gene Name
| GAPDH |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in glyceraldehyde-3-phosphate dehydrogenase activity |
| Specific Function
| Has both glyceraldehyde-3-phosphate dehydrogenase and nitrosylase activities, thereby playing a role in glycolysis and nuclear functions, respectively. Participates in nuclear events including transcription, RNA transport, DNA replication and apoptosis. Nuclear functions are probably due to the nitrosylase activity that mediates cysteine S-nitrosylation of nuclear target proteins such as SIRT1, HDAC2 and PRKDC. Modulates the organization and assembly of the cytoskeleton. Facilitates the CHP1-dependent microtubule and membrane associations through its ability to stimulate the binding of CHP1 to microtubules (By similarity). Glyceraldehyde-3-phosphate dehydrogenase is a key enzyme in glycolysis that catalyzes the first step of the pathway by converting D-glyceraldehyde 3-phosphate (G3P) into 3-phospho-D-glyceroyl phosphate. Component of the GAIT (gamma interferon-activated inhibitor of translation) complex which mediates interferon-gamma-induced transcript-selective translation inhibition in inflammation processes. Upon interferon-gamma treatment assembles into the GAIT complex which binds to stem loop-containing GAIT elements in the 3'-UTR of diverse inflammatory mRNAs (such as ceruplasmin) and suppresses their translation.
|
| GO Classification
|
| Biological Process |
| small molecule metabolic process |
| neuron apoptotic process |
| peptidyl-cysteine S-trans-nitrosylation |
| protein stabilization |
| glycolysis |
| negative regulation of translation |
| gluconeogenesis |
| cellular response to interferon-gamma |
| microtubule cytoskeleton organization |
| Cellular Component |
| plasma membrane |
| perinuclear region of cytoplasm |
| nucleus |
| lipid particle |
| ribonucleoprotein complex |
| cytosol |
| microtubule cytoskeleton |
| extracellular vesicular exosome |
| Function |
| binding |
| nucleotide binding |
| catalytic activity |
| nad or nadh binding |
| glyceraldehyde-3-phosphate dehydrogenase activity |
| oxidoreductase activity |
| oxidoreductase activity, acting on the aldehyde or oxo group of donors |
| oxidoreductase activity, acting on the aldehyde or oxo group of donors, nad or nadp as acceptor |
| Molecular Function |
| peptidyl-cysteine S-nitrosylase activity |
| microtubule binding |
| NAD binding |
| NADP binding |
| glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity |
| Process |
| metabolic process |
| small molecule metabolic process |
| alcohol metabolic process |
| monosaccharide metabolic process |
| hexose metabolic process |
| glucose metabolic process |
| oxidation reduction |
|
| Cellular Location
|
- Cytoplasm
- Cytoplasm
- perinuclear region
- Membrane
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Gluconeogenesis | 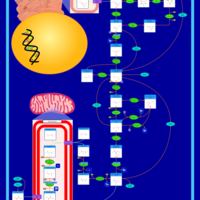 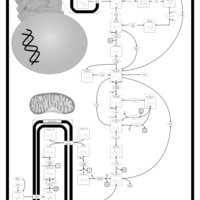 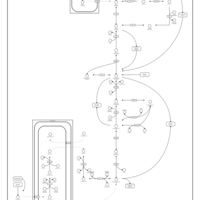 |  | | Glycerol Phosphate Shuttle | 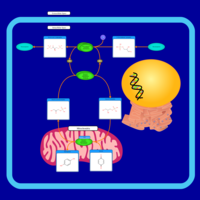 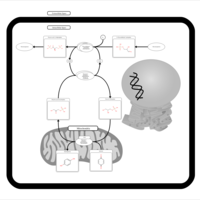  | Not Available | | Glycolysis | 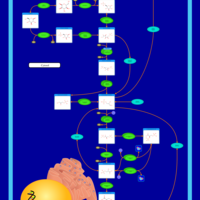 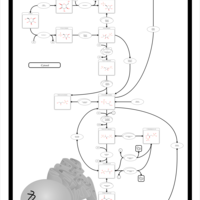 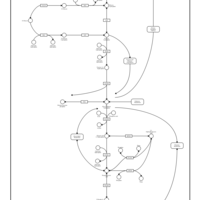 |  | | Mitochondrial Electron Transport Chain | 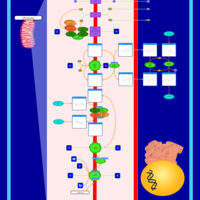 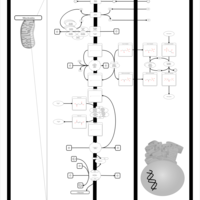 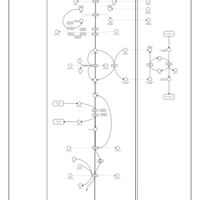 |  | | Glycolysis / Gluconeogenesis | Not Available |  |
|
| Gene Properties |
|---|
| Chromosome Location
| 12 |
| Locus
| 12p13 |
| SNPs
| GAPDH |
| Gene Sequence
|
>1008 bp
ATGGGGAAGGTGAAGGTCGGAGTCAACGGATTTGGTCGTATTGGGCGCCTGGTCACCAGG
GCTGCTTTTAACTCTGGTAAAGTGGATATTGTTGCCATCAATGACCCCTTCATTGACCTC
AACTACATGGTTTACATGTTCCAATATGATTCCACCCATGGCAAATTCCATGGCACCGTC
AAGGCTGAGAACGGGAAGCTTGTCATCAATGGAAATCCCATCACCATCTTCCAGGAGCGA
GATCCCTCCAAAATCAAGTGGGGCGATGCTGGCGCTGAGTACGTCGTGGAGTCCACTGGC
GTCTTCACCACCATGGAGAAGGCTGGGGCTCATTTGCAGGGGGGAGCCAAAAGGGTCATC
ATCTCTGCCCCCTCTGCTGATGCCCCCATGTTCGTCATGGGTGTGAACCATGAGAAGTAT
GACAACAGCCTCAAGATCATCAGCAATGCCTCCTGCACCACCAACTGCTTAGCACCCCTG
GCCAAGGTCATCCATGACAACTTTGGTATCGTGGAAGGACTCATGACCACAGTCCATGCC
ATCACTGCCACCCAGAAGACTGTGGATGGCCCCTCCGGGAAACTGTGGCGTGATGGCCGC
GGGGCTCTCCAGAACATCATCCCTGCCTCTACTGGCGCTGCCAAGGCTGTGGGCAAGGTC
ATCCCTGAGCTGAACGGGAAGCTCACTGGCATGGCCTTCCGTGTCCCCACTGCCAACGTG
TCAGTGGTGGACCTGACCTGCCGTCTAGAAAAACCTGCCAAATATGATGACATCAAGAAG
GTGGTGAAGCAGGCGTCGGAGGGCCCCCTCAAGGGCATCCTGGGCTACACTGAGCACCAG
GTGGTCTCCTCTGACTTCAACAGCGACACCCACTCCTCCACCTTTGACGCTGGGGCTGGC
ATTGCCCTCAACGACCACTTTGTCAAGCTCATTTCCTGGTATGACAACGAATTTGGCTAC
AGCAACAGGGTGGTGGACCTCATGGCCCACATGGCCTCCAAGGAGTAA
|
| Protein Properties |
|---|
| Number of Residues
| 335 |
| Molecular Weight
| 31547.76 |
| Theoretical pI
| 7.604 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Glyceraldehyde-3-phosphate dehydrogenase
MGKVKVGVNGFGRIGRLVTRAAFNSGKVDIVAINDPFIDLNYMVYMFQYDSTHGKFHGTV
KAENGKLVINGNPITIFQERDPSKIKWGDAGAEYVVESTGVFTTMEKAGAHLQGGAKRVI
ISAPSADAPMFVMGVNHEKYDNSLKIISNASCTTNCLAPLAKVIHDNFGIVEGLMTTVHA
ITATQKTVDGPSGKLWRDGRGALQNIIPASTGAAKAVGKVIPELNGKLTGMAFRVPTANV
SVVDLTCRLEKPAKYDDIKKVVKQASEGPLKGILGYTEHQVVSSDFNSDTHSSTFDAGAG
IALNDHFVKLISWYDNEFGYSNRVVDLMAHMASKE
|
| External Links |
|---|
| GenBank ID Protein
| 21104392 |
| UniProtKB/Swiss-Prot ID
| P04406 |
| UniProtKB/Swiss-Prot Entry Name
| G3P_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AB062273 |
| GeneCard ID
| GAPDH |
| GenAtlas ID
| GAPDH |
| HGNC ID
| HGNC:4141 |
| References |
|---|
| General References
| Not Available |

