| Identification |
|---|
| HMDB Protein ID
| CDBP00690 |
| Secondary Accession Numbers
| Not Available |
| Name
| Dihydropyrimidine dehydrogenase [NADP(+)] |
| Description
| Not Available |
| Synonyms
|
- DHPDHase
- DPD
- Dihydrothymine dehydrogenase
- Dihydrouracil dehydrogenase
|
| Gene Name
| DPYD |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in electron carrier activity |
| Specific Function
| Involved in pyrimidine base degradation. Catalyzes the reduction of uracil and thymine. Also involved the degradation of the chemotherapeutic drug 5-fluorouracil.
|
| GO Classification
|
| Biological Process |
| beta-alanine biosynthetic process |
| purine nucleobase catabolic process |
| thymidine catabolic process |
| thymine catabolic process |
| UMP biosynthetic process |
| uracil catabolic process |
| Cellular Component |
| cytosol |
| Component |
| cell part |
| intracellular part |
| cytoplasm |
| Function |
| dihydroorotate oxidase activity |
| oxidoreductase activity, acting on the ch-ch group of donors, quinone or related compound as acceptor |
| dihydroorotate dehydrogenase activity |
| oxidoreductase activity |
| oxidoreductase activity, acting on the ch-ch group of donors |
| binding |
| catalytic activity |
| electron carrier activity |
| metal cluster binding |
| iron-sulfur cluster binding |
| Molecular Function |
| metal ion binding |
| NADP binding |
| oxidoreductase activity |
| 4 iron, 4 sulfur cluster binding |
| protein homodimerization activity |
| dihydroorotate oxidase activity |
| dihydropyrimidine dehydrogenase (NADP+) activity |
| iron-sulfur cluster binding |
| flavin adenine dinucleotide binding |
| Process |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| nucleobase, nucleoside and nucleotide metabolic process |
| nucleoside phosphate metabolic process |
| nucleotide metabolic process |
| cellular aromatic compound metabolic process |
| cellular metabolic process |
| nucleobase metabolic process |
| pyrimidine base metabolic process |
| pyrimidine base biosynthetic process |
| 'de novo' pyrimidine base biosynthetic process |
| pyrimidine nucleoside monophosphate biosynthetic process |
| pyrimidine ribonucleoside monophosphate biosynthetic process |
| ump biosynthetic process |
| oxidation reduction |
| metabolic process |
| pyrimidine nucleotide metabolic process |
| nitrogen compound metabolic process |
| pyrimidine nucleotide biosynthetic process |
| cellular nitrogen compound metabolic process |
|
| Cellular Location
|
- Cytoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | beta-alanine biosynthesis | Not Available | Not Available | | Drug metabolism - other enzymes | Not Available | 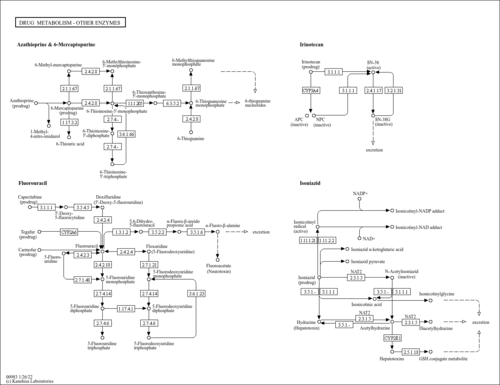 | | Beta-Alanine Metabolism |    |  | | GABA-Transaminase Deficiency |    | Not Available | | Ureidopropionase Deficiency |    | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 1 |
| Locus
| 1p22 |
| SNPs
| DPYD |
| Gene Sequence
|
>3078 bp
ATGGCCCCTGTGCTCAGTAAGGACTCGGCGGACATCGAGAGTATCCTGGCTTTAAATCCT
CGAACACAAACTCATGCAACTCTGTGTTCCACTTCGGCCAAGAAATTAGACAAGAAACAT
TGGAAAAGAAATCCTGATAAGAACTGCTTTAATTGTGAGAAGCTGGAGAATAATTTTGAT
GACATCAAGCACACGACTCTTGGTGAGCGAGGAGCTCTCCGAGAAGCAATGAGATGCCTG
AAATGTGCAGATGCCCCGTGTCAGAAGAGCTGTCCAACTAATCTTGATATTAAATCATTC
ATCACAAGTATTGCAAACAAGAACTATTATGGAGCTGCTAAGATGATATTTTCTGACAAC
CCACTTGGTCTGACTTGTGGAATGGTATGTCCAACCTCTGATCTTTGTGTAGGTGGATGC
AATTTATATGCCACTGAAGAGGGACCCATTAATATTGGTGGATTGCAGCAATTTGCTACT
GAGGTATTCAAAGCAATGAGTATCCCACAGATCAGAAATCCTTCGCTGCCTCCCCCAGAA
AAAATGTCTGAAGCCTATTCTGCAAAGATTGCTCTTTTTGGTGCTGGGCCTGCAAGTATA
AGTTGTGCTTCCTTTTTGGCTCGATTGGGGTACTCTGACATCACTATATTTGAAAAACAA
GAATATGTTGGTGGTTTAAGTACTTCTGAAATTCCTCAGTTCCGGCTGCCGTATGATGTA
GTGAATTTTGAGATTGAGCTAATGAAGGACCTTGGTGTAAAGATAATTTGCGGTAAAAGC
CTTTCAGTGAATGAAATGACTCTTAGCACTTTGAAAGAAAAAGGCTACAAAGCTGCTTTC
ATTGGAATAGGTTTGCCAGAACCCAATAAAGATGCCATCTTCCAAGGCCTGACGCAGGAC
CAGGGGTTTTATACATCCAAAGACTTTTTGCCACTTGTAGCCAAAGGCAGTAAAGCAGGA
ATGTGCGCCTGTCACTCTCCATTGCCATCGATACGGGGAGTCGTGATCGTACTTGGAGCT
GGAGACACTGCCTTTGACTGTGCAACATCTGCTCTACGTTGTGGAGCTCGCCGTGTGTTC
ATCGTCTTCAGAAAAGGCTTTGTTAATATAAGAGCTGTCCCTGAGGAGATGGAACTTGCT
AAGGAAGAAAAGTGTGAATTTCTGCCATTCCTGTCCCCACGGAAGGTTATAGTAAAAGGT
GGGAGAATTGTTGCTATGCAGTTTGTTCGGACAGAGCAAGATGAAACTGGAAAATGGAAT
GAAGATGAAGATCAGATGGTCCATCTGAAAGCCGATGTGGTCATCAGTGCCTTTGGTTCA
GTTCTGAGTGATCCTAAAGTAAAAGAAGCCTTGAGCCCTATAAAATTTAACAGATGGGGT
CTCCCAGAAGTAGATCCAGAAACTATGCAAACTAGTGAAGCATGGGTATTTGCAGGTGGT
GATGTCGTTGGTTTGGCTAACACTACAGTGGAATCGGTGAATGATGGAAAGCAAGCTTCT
TGGTACATTCACAAATACGTACAGTCACAATATGGAGCTTCCGTTTCTGCCAAGCCTGAA
CTACCCCTCTTTTACACTCCTATTGATCTGGTGGACATTAGTGTAGAAATGGCCGGATTG
AAGTTTATAAATCCTTTTGGTCTTGCTAGCGCAACTCCAGCCACCAGCACATCAATGATT
CGAAGAGCTTTTGAAGCTGGATGGGGTTTTGCCCTCACCAAAACTTTCTCTCTTGATAAG
GACATTGTGACAAATGTTTCCCCCAGAATCATCCGGGGAACCACCTCTGGCCCCATGTAT
GGCCCTGGACAAAGCTCCTTTCTGAATATTGAGCTCATCAGTGAGAAAACGGCTGCATAT
TGGTGTCAAAGTGTCACTGAACTAAAGGCTGACTTTCCAGACAACATTGTGATTGCTAGC
ATTATGTGCAGTTACAATAAAAATGACTGGACGGAACTTGCCAAGAAGTCTGAGGATTCT
GGAGCAGATGCCCTGGAGTTAAATTTATCATGTCCACATGGCATGGGAGAAAGAGGAATG
GGCCTGGCCTGTGGGCAGGATCCAGAGCTGGTGCGGAACATCTGCCGCTGGGTTAGGCAA
GCTGTTCAGATTCCTTTTTTTGCCAAGCTGACCCCAAATGTCACTGATATTGTGAGCATC
GCAAGAGCTGCAAAGGAAGGTGGTGCCAATGGCGTTACAGCCACCAACACTGTCTCAGGT
CTGATGGGATTAAAATCTGATGGCACACCTTGGCCAGCAGTGGGGATTGCAAAGCGAACT
ACATATGGAGGAGTGTCTGGGACAGCAATCAGACCTATTGCTTTGAGAGCTGTGACCTCC
ATTGCTCGTGCTCTGCCTGGATTTCCCATTTTGGCTACTGGTGGAATTGACTCTGCTGAA
AGTGGTCTTCAGTTTCTCCATAGTGGTGCTTCCGTCCTCCAGGTATGCAGTGCCATTCAG
AATCAGGATTTCACTGTGATCGAAGACTACTGCACTGGCCTCAAAGCCCTGCTTTATCTG
AAAAGCATTGAAGAACTACAAGACTGGGATGGACAGAGTCCAGCTACTGTGAGTCACCAG
AAAGGGAAACCAGTTCCACGTATAGCTGAACTCATGGACAAGAAACTGCCAAGTTTTGGA
CCTTATCTGGAACAGCGCAAGAAAATCATAGCAGAAAACAAGATTAGACTGAAAGAACAA
AATGTAGCTTTTTCACCACTTAAGAGAAACTGTTTTATCCCCAAAAGGCCTATTCCTACC
ATCAAGGATGTAATAGGAAAAGCACTGCAGTACCTTGGAACATTTGGTGAATTGAGCAAC
GTAGAGCAAGTTGTGGCTATGATTGATGAAGAAATGTGTATCAACTGTGGTAAATGCTAC
ATGACCTGTAATGATTCTGGCTACCAGGCTATACAGTTTGATCCAGAAACCCACCTGCCC
ACCATAACCGACACTTGTACAGGCTGTACTCTGTGTCTCAGTGTTTGCCCTATTGTCGAC
TGCATCAAAATGGTTTCCAGGACAACACCTTATGAACCAAAGAGAGGCGTACCCTTATCT
GTGAATCCGGTGTGTTAA
|
| Protein Properties |
|---|
| Number of Residues
| 1025 |
| Molecular Weight
| 111400.32 |
| Theoretical pI
| 7.05 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Dihydropyrimidine dehydrogenase [NADP+]
MAPVLSKDSADIESILALNPRTQTHATLCSTSAKKLDKKHWKRNPDKNCFNCEKLENNFD
DIKHTTLGERGALREAMRCLKCADAPCQKSCPTNLDIKSFITSIANKNYYGAAKMIFSDN
PLGLTCGMVCPTSDLCVGGCNLYATEEGPINIGGLQQFATEVFKAMSIPQIRNPSLPPPE
KMSEAYSAKIALFGAGPASISCASFLARLGYSDITIFEKQEYVGGLSTSEIPQFRLPYDV
VNFEIELMKDLGVKIICGKSLSVNEMTLSTLKEKGYKAAFIGIGLPEPNKDAIFQGLTQD
QGFYTSKDFLPLVAKGSKAGMCACHSPLPSIRGVVIVLGAGDTAFDCATSALRCGARRVF
IVFRKGFVNIRAVPEEMELAKEEKCEFLPFLSPRKVIVKGGRIVAMQFVRTEQDETGKWN
EDEDQMVHLKADVVISAFGSVLSDPKVKEALSPIKFNRWGLPEVDPETMQTSEAWVFAGG
DVVGLANTTVESVNDGKQASWYIHKYVQSQYGASVSAKPELPLFYTPIDLVDISVEMAGL
KFINPFGLASATPATSTSMIRRAFEAGWGFALTKTFSLDKDIVTNVSPRIIRGTTSGPMY
GPGQSSFLNIELISEKTAAYWCQSVTELKADFPDNIVIASIMCSYNKNDWTELAKKSEDS
GADALELNLSCPHGMGERGMGLACGQDPELVRNICRWVRQAVQIPFFAKLTPNVTDIVSI
ARAAKEGGANGVTATNTVSGLMGLKSDGTPWPAVGIAKRTTYGGVSGTAIRPIALRAVTS
IARALPGFPILATGGIDSAESGLQFLHSGASVLQVCSAIQNQDFTVIEDYCTGLKALLYL
KSIEELQDWDGQSPATVSHQKGKPVPRIAELMDKKLPSFGPYLEQRKKIIAENKIRLKEQ
NVAFSPLKRNCFIPKRPIPTIKDVIGKALQYLGTFGELSNVEQVVAMIDEEMCINCGKCY
MTCNDSGYQAIQFDPETHLPTITDTCTGCTLCLSVCPIVDCIKMVSRTTPYEPKRGVPLS
VNPVC
|
| External Links |
|---|
| GenBank ID Protein
| 6729338 |
| UniProtKB/Swiss-Prot ID
| Q12882 |
| UniProtKB/Swiss-Prot Entry Name
| DPYD_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| AB003063 |
| GeneCard ID
| DPYD |
| GenAtlas ID
| DPYD |
| HGNC ID
| HGNC:3012 |
| References |
|---|
| General References
| Not Available |

