| Identification |
|---|
| HMDB Protein ID
| CDBP00951 |
| Secondary Accession Numbers
| Not Available |
| Name
| Leukotriene-B(4) omega-hydroxylase 1 |
| Description
| Not Available |
| Synonyms
|
- CYPIVF2
- Cytochrome P450 4F2
- Cytochrome P450-LTB-omega
- Leukotriene-B(4) 20-monooxygenase 1
|
| Gene Name
| CYP4F2 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in monooxygenase activity |
| Specific Function
| Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics.
|
| GO Classification
|
| Biological Process |
| very long-chain fatty acid metabolic process |
| drug metabolic process |
| xenobiotic metabolic process |
| renal water homeostasis |
| sodium ion homeostasis |
| epoxygenase P450 pathway |
| positive regulation of icosanoid secretion |
| pressure natriuresis |
| negative regulation of blood coagulation |
| negative regulation of icosanoid secretion |
| vitamin E metabolic process |
| vitamin K biosynthetic process |
| Cellular Component |
| endoplasmic reticulum membrane |
| apical plasma membrane |
| Function |
| metal ion binding |
| binding |
| catalytic activity |
| transition metal ion binding |
| electron carrier activity |
| iron ion binding |
| monooxygenase activity |
| heme binding |
| oxidoreductase activity |
| ion binding |
| cation binding |
| Molecular Function |
| alpha-tocopherol omega-hydroxylase activity |
| tocotrienol omega-hydroxylase activity |
| vitamin-K-epoxide reductase (warfarin-sensitive) activity |
| electron carrier activity |
| iron ion binding |
| heme binding |
| alkane 1-monooxygenase activity |
| arachidonic acid epoxygenase activity |
| arachidonic acid omega-hydroxylase activity |
| leukotriene-B4 20-monooxygenase activity |
| Process |
| metabolic process |
| oxidation reduction |
|
| Cellular Location
|
- Peripheral membrane protein
- Peripheral membrane protein
- Endoplasmic reticulum membrane
- Microsome membrane
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Arachidonic Acid Metabolism | 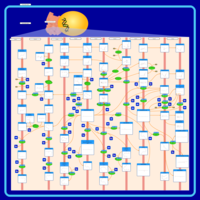  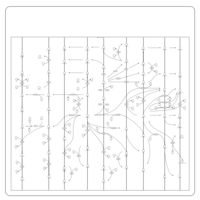 |  | | Leukotriene C4 Synthesis Deficiency | 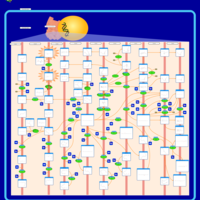 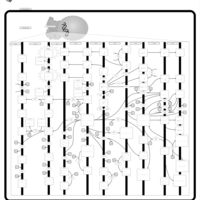  | Not Available | | Piroxicam Action Pathway |    | Not Available | | Acetylsalicylic Acid Action Pathway |  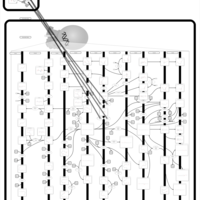 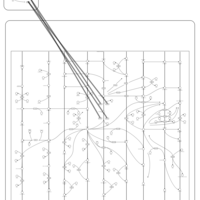 | Not Available | | Etodolac Action Pathway |  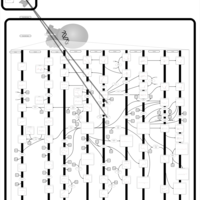  | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 19 |
| Locus
| 19p13.12 |
| SNPs
| CYP4F2 |
| Gene Sequence
|
>1563 bp
ATGTCCCAGCTGAGCCTGTCCTGGCTGGGCCTCGGGCCAGTGGCAGCATCCCCTTGGCTG
CTCCTCCTGCTGGTCGGGGCCTCCTGGCTCCTGGCCCATGTCCTGGCCTGGACCTACGCC
TTCTATGACAACTGCCGCCGCCTTCGGTGTTTCCCACAACCCCCGAGACGGAACTGGTTT
TGGGGACACCAGGGCATGGTCAACCCCACAGAGGAGGGCATGAGAGTTCTGACTCAGCTG
GTGGCCACCTACCCCCAGGGCTTTAAGGTCTGGATGGGACCCATCTCCCCCCTCCTCAGT
TTGTGCCACCCCGACATCATCCGGTCTGTCATCAACGCCTCAGCTGCCATTGCACCAAAG
GACAAGTTCTTCTACAGCTTCCTGGAGCCCTGGCTGGGGGATGGGCTCCTGCTGAGTGCT
GGTGACAAGTGGAGCCGCCACCGTCGGATGCTGACGCCTGCCTTCCATTTCAACATCCTG
AAGCCCTATATGAAGATTTTCAATGAGAGTGTGAACATCATGCACGCCAAGTGGCAGCTC
CTGGCCTCAGAGGGTAGTGCCTGTTTGGATATGTTTGAGCACATCAGCCTCATGACCTTG
GACAGTCTACAGAAATGTGTCTTCAGCTTTGACAGCCATTGTCAGGAGAAACCCAGTGAA
TATATTGCCGCCATCTTGGAGCTCAGTGCCCTTGTATCAAAAAGACACCATGAGATCCTC
CTGCATATTGACTTCCTGTATTATCTCACCCCTGATGGGCAGCGTTTCCGCAGGGCCTGC
CGCCTGGTGCACGACTTCACAGATGCCGTCATCCAGGAGCGGCGCCGCACTCTCCCTAGC
CAGGGTGTTGATGACTTCCTCCAAGCCAAGGCCAAATCCAAGACTTTGGACTTCATTGAT
GTACTCCTGCTGAGCAAGGATGAAGACGGGAAGAAGTTATCTGATGAGGACATAAGAGCA
GAAGCTGACACCTTTATGTTTGAGGGCCATGACACCACGGCCAGTGGTCTCTCCTGGGTC
CTGTACCACCTTGCAAAGCACCCAGAATACCAGGAGCGCTGCCGGCAGGAGGTGCAAGAA
CTTCTGAAGGACCGTGAGCCTAAAGAGATTGAATGGGACGACCTGGCCCATTTGCCCTTC
CTGACCATGTGCATGAAGGAGAGCCTGCGGCTGCATCCCCCAGTCCCGGTCATCTCCCGC
CATGTCACCCAGGACATTGTGCTCCCAGACGGCCGGGTCATCCCCAAAGGCATTATCTGC
CTCATCAGTGTTTTCGGAACCCATCACAACCCAGCTATGTGGCCGGACCCTGAGGTCTAC
GACCCCTTTCGCTTTGACCCAGAGAACATCAAGGAGAGGTCACCTCTGGCTTTTATTCCC
TTCTCGGCAGGGCCCAGGAACTGCATCGGGCAGACGTTCGCGATGGCGGAGATGAAGGTG
GTCCTGGCGCTCACGCTGCTGCGCTTCCGCGTCCTGCCTGACCACACCGAGCCCCGCAGG
AAGCCGGAGCTGGTCCTGCGCGCAGAGGGCGGACTTTGGCTGCGGGTGGAGCCCCTGAGC
TGA
|
| Protein Properties |
|---|
| Number of Residues
| 520 |
| Molecular Weight
| 59852.825 |
| Theoretical pI
| 7.075 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Leukotriene-B(4) omega-hydroxylase 1
MSQLSLSWLGLWPVAASPWLLLLLVGASWLLAHVLAWTYAFYDNCRRLRCFPQPPRRNWF
WGHQGMVNPTEEGMRVLTQLVATYPQGFKVWMGPISPLLSLCHPDIIRSVINASAAIAPK
DKFFYSFLEPWLGDGLLLSAGDKWSRHRRMLTPAFHFNILKPYMKIFNESVNIMHAKWQL
LASEGSACLDMFEHISLMTLDSLQKCVFSFDSHCQEKPSEYIAAILELSALVSKRHHEIL
LHIDFLYYLTPDGQRFRRACRLVHDFTDAVIQERRRTLPSQGVDDFLQAKAKSKTLDFID
VLLLSKDEDGKKLSDEDIRAEADTFMFEGHDTTASGLSWVLYHLAKHPEYQERCRQEVQE
LLKDREPKEIEWDDLAHLPFLTMCMKESLRLHPPVPVISRHVTQDIVLPDGRVIPKGIIC
LISVFGTHHNPAVWPDPEVYDPFRFDPENIKERSPLAFIPFSAGPRNCIGQTFAMAEMKV
VLALTLLRFRVLPDHTEPRRKPELVLRAEGGLWLRVEPLS
|
| External Links |
|---|
| GenBank ID Protein
| 4519535 |
| UniProtKB/Swiss-Prot ID
| P78329 |
| UniProtKB/Swiss-Prot Entry Name
| CP4F2_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| AB015306 |
| GeneCard ID
| CYP4F2 |
| GenAtlas ID
| CYP4F2 |
| HGNC ID
| HGNC:2645 |
| References |
|---|
| General References
| Not Available |
