| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:16:52 UTC |
|---|
| Updated at | 2020-11-18 16:39:28 UTC |
|---|
| CannabisDB ID | CDB005175 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Heme |
|---|
| Description | Heme is also known as protoheme or [fe(ppix)]. Heme is a weakly acidic compound (based on its pKa). Heme exists in all living organisms, ranging from bacteria to humans. In humans, heme is involved in ibuprofen action pathway. Outside of the human body, Heme has been detected, but not quantified in, several different foods, such as triticales, chicories, quinces, black mulberries, and japanese walnuts. This could make heme a potential biomarker for the consumption of these foods. Heme in which the iron has oxidation state +2. Heme is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| [3,7,12,17-Tetramethyl-8,13-divinylporphyrin-2,18-dipropanoato(2-)]iron(II) | ChEBI | | [Fe(ppix)] | ChEBI | | Fe(ppix) | ChEBI | | Ferroprotoheme | ChEBI | | Ferroprotoporphyrin IX | ChEBI | | Ferrous protoheme | ChEBI | | Ferrous protoheme IX | ChEBI | | Haem | ChEBI | | Iron(II) protoporphyrin IX | ChEBI | | Protoferroheme | ChEBI | | Protoheme | ChEBI | | Heme b | Kegg | | Protoheme IX | Kegg | | Heme | ChEBI |
|
|---|
| Chemical Formula | C34H32FeN4O4 |
|---|
| Average Molecular Weight | 616.49 |
|---|
| Monoisotopic Molecular Weight | 616.1773 |
|---|
| IUPAC Name | 4,20-bis(2-carboxyethyl)-10,15-diethenyl-5,9,14,19-tetramethyl-2lambda5,22,23lambda5,25-tetraaza-1-ferraoctacyclo[11.9.1.1^{1,8}.1^{3,21}.0^{2,6}.0^{16,23}.0^{18,22}.0^{11,25}]pentacosa-2,4,6,8,10,12,14,16(23),17,19,21(24)-undecaene-2,23-bis(ylium)-1,1-diuide |
|---|
| Traditional Name | 4,20-bis(2-carboxyethyl)-10,15-diethenyl-5,9,14,19-tetramethyl-2lambda5,22,23lambda5,25-tetraaza-1-ferraoctacyclo[11.9.1.1^{1,8}.1^{3,21}.0^{2,6}.0^{16,23}.0^{18,22}.0^{11,25}]pentacosa-2,4,6,8,10,12,14,16(23),17,19,21(24)-undecaene-2,23-bis(ylium)-1,1-diuide |
|---|
| CAS Registry Number | 14875-96-8 |
|---|
| SMILES | CC1=C(CCC(O)=O)C2=CC3=[N+]4C(=CC5=C(C)C(C=C)=C6C=C7C(C)=C(C=C)C8=[N+]7[Fe--]4(N2C1=C8)N56)C(C)=C3CCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C34H34N4O4.Fe/c1-7-21-17(3)25-13-26-19(5)23(9-11-33(39)40)31(37-26)16-32-24(10-12-34(41)42)20(6)28(38-32)15-30-22(8-2)18(4)27(36-30)14-29(21)35-25;/h7-8,13-16H,1-2,9-12H2,3-6H3,(H4,35,36,37,38,39,40,41,42);/q;+2/p-2/b25-13-,26-13-,27-14-,28-15-,29-14-,30-15-,31-16-,32-16-; |
|---|
| InChI Key | KABFMIBPWCXCRK-RGGAHWMASA-L |
|---|
| Chemical Taxonomy |
|---|
| Classification | Not classified |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Heme, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Heme, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Heme, TMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Heme, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Heme, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Heme, TBDMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Heme, "Heme,1TMS,#1" TMS, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-066r-0000095000-501c4a776d38d61ab89a | 2019-02-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0gi4-0000092000-64c21d0a0f8daa069fb9 | 2019-02-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03gs-4000590000-15136ce22e51bdc57377 | 2019-02-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000049000-69ee302652cebc40f13b | 2019-02-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00r2-1000091000-d178bb83079a203a48dc | 2019-02-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-1000090000-f6e437751fbc08f12d1f | 2019-02-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014j-0000059000-0ab427d87ba10992d498 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0000098000-650f7d2f023028ff2319 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-06fr-0000090000-2dff1da74eb12a246237 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-0000079000-990502ea40621d9cad46 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05ns-0000091000-3547d5c809ccf8987200 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0cdr-0000092000-e55d1ab39076367d5b9e | 2021-09-25 | View Spectrum |
|
|---|
| NMR | Not Available |
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Porphyrin Metabolism | 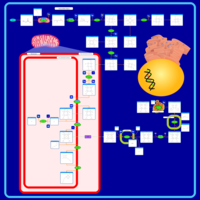 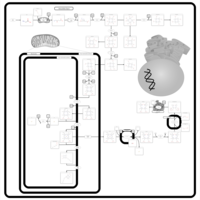 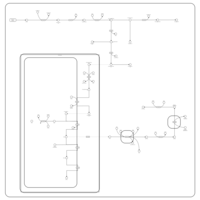 |  | | Degradation of Superoxides | 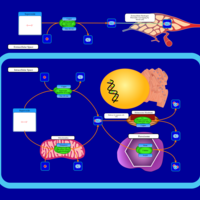   | Not Available | | Ethanol Degradation | 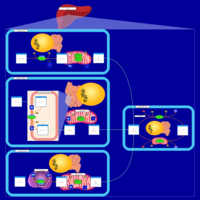 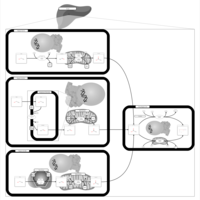  | Not Available | | Disulfiram Action Pathway | 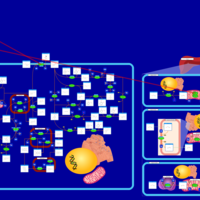 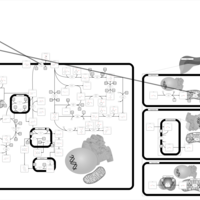 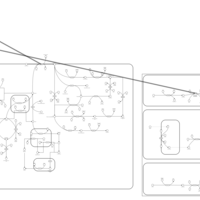 | Not Available | | Sulfate/Sulfite Metabolism | 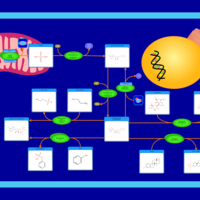   |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| 5-aminolevulinate synthase, erythroid-specific, mitochondrial | ALAS2 | Xp11.21 | P22557 | details | | 5-aminolevulinate synthase, nonspecific, mitochondrial | ALAS1 | 3p21.1 | P13196 | details | | Succinate dehydrogenase cytochrome b560 subunit, mitochondrial | SDHC | 1q23.3 | Q99643 | details | | Aldehyde oxidase | AOX1 | 2q33 | Q06278 | details | | Heme oxygenase 2 | HMOX2 | 16p13.3 | P30519 | details | | Succinate dehydrogenase [ubiquinone] cytochrome b small subunit, mitochondrial | SDHD | 11q23 | O14521 | details | | NADPH--cytochrome P450 reductase | POR | 7q11.2 | P16435 | details | | Heme oxygenase 1 | HMOX1 | 22q13.1 | P09601 | details | | Uroporphyrinogen decarboxylase | UROD | 1p34 | P06132 | details | | Coproporphyrinogen-III oxidase, mitochondrial | CPOX | 3q12 | P36551 | details | | Lanosterol 14-alpha demethylase | CYP51A1 | 7q21.2 | Q16850 | details | | Thyroid peroxidase | TPO | 2p25 | P07202 | details | | Lactoperoxidase | LPO | 17q23.1 | P22079 | details | | Myeloperoxidase | MPO | 17q23.1 | P05164 | details | | Eosinophil peroxidase | EPX | 17q23.1 | P11678 | details | | Uroporphyrinogen-III synthase | UROS | 10q25.2-q26.3 | P10746 | details | | Porphobilinogen deaminase | HMBS | 11q23.3 | P08397 | details | | Indoleamine 2,3-dioxygenase 1 | IDO1 | 8p12-p11 | P14902 | details | | Cholesterol 7-alpha-monooxygenase | CYP7A1 | 8q11-q12 | P22680 | details | | Biliverdin reductase A | BLVRA | 7p13 | P53004 | details | | Flavin reductase (NADPH) | BLVRB | 19q13.1-q13.2 | P30043 | details | | Cystathionine beta-synthase | CBS | 21q22.3 | P35520 | details | | Nitric oxide synthase, inducible | NOS2 | 17q11.2-q12 | P35228 | details | | Nitric oxide synthase, brain | NOS1 | 12q24.2-q24.31 | P29475 | details | | Nitric oxide synthase, endothelial | NOS3 | 7q36 | P29474 | details | | Succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial | SUCLA2 | 13q12.2-q13.3 | Q9P2R7 | details | | Prostaglandin G/H synthase 2 | PTGS2 | 1q25.2-q25.3 | P35354 | details | | Prostaglandin G/H synthase 1 | PTGS1 | 9q32-q33.3 | P23219 | details | | Leukotriene-B(4) omega-hydroxylase 1 | CYP4F2 | 19p13.12 | P78329 | details | | Leukotriene-B(4) omega-hydroxylase 2 | CYP4F3 | 19p13.2 | Q08477 | details | | Protoporphyrinogen oxidase | PPOX | 1q22 | P50336 | details | | Sterol 26-hydroxylase, mitochondrial | CYP27A1 | 2q33-qter | Q02318 | details | | 25-hydroxyvitamin D-1 alpha hydroxylase, mitochondrial | CYP27B1 | 12q13.1-q13.3 | O15528 | details | | Steroid 17-alpha-hydroxylase/17,20 lyase | CYP17A1 | 10q24.3 | P05093 | details | | Sulfite oxidase, mitochondrial | SUOX | 12q13.2 | P51687 | details | | Ferrochelatase, mitochondrial | FECH | 18q21.3 | P22830 | details | | Delta-aminolevulinic acid dehydratase | ALAD | 9q33.1 | P13716 | details | | Guanylate cyclase soluble subunit beta-1 | GUCY1B3 | 4q31.3-q33 | Q02153 | details | | Guanylate cyclase soluble subunit beta-2 | GUCY1B2 | | O75343 | details | | Prostacyclin synthase | PTGIS | 20q13.13 | Q16647 | details | | Catalase | CAT | 11p13 | P04040 | details | | Thromboxane-A synthase | TBXAS1 | 7q34-q35 | P24557 | details | | 25-hydroxycholesterol 7-alpha-hydroxylase | CYP7B1 | 8q21.3 | O75881 | details | | Cytochrome c oxidase subunit 1 | MT-CO1 | | P00395 | details | | Protoheme IX farnesyltransferase, mitochondrial | COX10 | | Q12887 | details | | Cytochrome c-type heme lyase | HCCS | Xp22.3 | P53701 | details | | 1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial | CYP24A1 | 20q13 | Q07973 | details | | Fatty acid desaturase 1 | FADS1 | 11q12.2-q13.1 | O60427 | details | | Vitamin D 25-hydroxylase | CYP2R1 | 11p15.2 | Q6VVX0 | details | | Cytochrome b reductase 1 | CYBRD1 | 2q31.1 | Q53TN4 | details | | Cytochrome c oxidase subunit 5A, mitochondrial | COX5A | 15q24.1 | P20674 | details | | Fatty acid desaturase 2 | FADS2 | 11q12.2 | O95864 | details | | Cholesterol 24-hydroxylase | CYP46A1 | 14q32.1 | Q9Y6A2 | details | | 24-hydroxycholesterol 7-alpha-hydroxylase | CYP39A1 | 6p21.1-p11.2 | Q9NYL5 | details | | 7-alpha-hydroxycholest-4-en-3-one 12-alpha-hydroxylase | CYP8B1 | 3p22.1 | Q9UNU6 | details | | Cytochrome b5 reductase 4 | CYB5R4 | 6pter-q22.33 | Q7L1T6 | details | | Indoleamine 2,3-dioxygenase 2 | IDO2 | 8p11.21 | Q6ZQW0 | details | | Cytochrome P450, family 1, subfamily A, polypeptide 1 | CYP1A1 | 15q24.1 | A0N0X8 | details | | NADPH oxidase 5 | NOX5 | 15q23 | Q96PH1 | details | | Metalloreductase STEAP3 | STEAP3 | 2q14.2 | Q658P3 | details | | NADPH oxidase 1 | NOX1 | | Q9Y5S8 | details | | NADPH oxidase 4 | NOX4 | 11q14.2-q21 | Q9NPH5 | details | | Metalloreductase STEAP1 | STEAP1 | 7q21 | Q9UHE8 | details | | Metalloreductase STEAP2 | STEAP2 | 7q21 | Q8NFT2 | details | | Metalloreductase STEAP4 | STEAP4 | 7q21.12 | Q687X5 | details | | Cytochrome c oxidase assembly protein COX15 homolog | COX15 | | Q7KZN9 | details | | Eukaryotic translation initiation factor 2-alpha kinase 1 | EIF2AK1 | 7p22 | Q9BQI3 | details | | Fatty acid 2-hydroxylase | FA2H | 16q23 | Q7L5A8 | details | | Fatty acid desaturase 2-like protein | | | A8MWK0 | details | | Fatty acid desaturase 3 | FADS3 | 11q12-q13.1 | Q9Y5Q0 | details | | Ferric-chelate reductase 1 | FRRS1 | 1p21.2 | Q6ZNA5 | details | | E3 ubiquitin-protein ligase HERC2 | HERC2 | 15q13 | O95714 | details | | Cytochrome P450, family 21, subfamily A, polypeptide 2 | CYP21A2 | 6p21.3 | Q08AG9 | details |
|
|---|
| Transporters | |
| Nitric oxide synthase, inducible | NOS2 | 17q11.2-q12 | P35228 | details | | Cytochrome b-c1 complex subunit 9 | UQCR10 | 22cen-q12.3 | Q9UDW1 | details | | Cytochrome b-c1 complex subunit 6, mitochondrial | UQCRH | 1p34.1 | P07919 | details | | Protein AMBP | AMBP | 9q32-q33 | P02760 | details | | Calcium-activated potassium channel subunit alpha-1 | KCNMA1 | 10q22.3 | Q12791 | details | | Proton-coupled folate transporter | SLC46A1 | 17q11.2 | Q96NT5 | details | | ATP-binding cassette sub-family B member 6, mitochondrial | ABCB6 | 2q36 | Q9NP58 | details | | ATP-binding cassette sub-family B member 7, mitochondrial | ABCB7 | | O75027 | details | | ATP-binding cassette sub-family B member 10, mitochondrial | ABCB10 | 1q42.13 | Q9NRK6 | details | | Heme transporter HRG1 | SLC48A1 | 12q13.11 | Q6P1K1 | details |
|
|---|
| Metal Bindings | |
| Succinate dehydrogenase cytochrome b560 subunit, mitochondrial | SDHC | 1q23.3 | Q99643 | details | | Aldehyde oxidase | AOX1 | 2q33 | Q06278 | details | | Heme oxygenase 2 | HMOX2 | 16p13.3 | P30519 | details | | Succinate dehydrogenase [ubiquinone] cytochrome b small subunit, mitochondrial | SDHD | 11q23 | O14521 | details | | NADPH--cytochrome P450 reductase | POR | 7q11.2 | P16435 | details | | Heme oxygenase 1 | HMOX1 | 22q13.1 | P09601 | details | | Lanosterol 14-alpha demethylase | CYP51A1 | 7q21.2 | Q16850 | details | | Thyroid peroxidase | TPO | 2p25 | P07202 | details | | Lactoperoxidase | LPO | 17q23.1 | P22079 | details | | Myeloperoxidase | MPO | 17q23.1 | P05164 | details | | Eosinophil peroxidase | EPX | 17q23.1 | P11678 | details | | Cytochrome P450 4A11 | CYP4A11 | 1p33 | Q02928 | details | | Indoleamine 2,3-dioxygenase 1 | IDO1 | 8p12-p11 | P14902 | details | | Cytochrome P450 11B1, mitochondrial | CYP11B1 | 8q21 | P15538 | details | | Cholesterol 7-alpha-monooxygenase | CYP7A1 | 8q11-q12 | P22680 | details | | Biliverdin reductase A | BLVRA | 7p13 | P53004 | details | | Cholesterol side-chain cleavage enzyme, mitochondrial | CYP11A1 | 15q23-q24 | P05108 | details | | Cystathionine beta-synthase | CBS | 21q22.3 | P35520 | details | | Nitric oxide synthase, inducible | NOS2 | 17q11.2-q12 | P35228 | details | | Nitric oxide synthase, brain | NOS1 | 12q24.2-q24.31 | P29475 | details | | Nitric oxide synthase, endothelial | NOS3 | 7q36 | P29474 | details | | Succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial | SUCLA2 | 13q12.2-q13.3 | Q9P2R7 | details | | Prostaglandin G/H synthase 2 | PTGS2 | 1q25.2-q25.3 | P35354 | details | | Prostaglandin G/H synthase 1 | PTGS1 | 9q32-q33.3 | P23219 | details | | Cytochrome c1, heme protein, mitochondrial | CYC1 | 8q24.3 | P08574 | details | | Leukotriene-B(4) omega-hydroxylase 1 | CYP4F2 | 19p13.12 | P78329 | details | | Leukotriene-B(4) omega-hydroxylase 2 | CYP4F3 | 19p13.2 | Q08477 | details | | Cytochrome P450 3A4 | CYP3A4 | 7q21.1 | P08684 | details | | Cytochrome P450 2C9 | CYP2C9 | 10q24 | P11712 | details | | Sterol 26-hydroxylase, mitochondrial | CYP27A1 | 2q33-qter | Q02318 | details | | Cytochrome P450 2C19 | CYP2C19 | 10q24 | P33261 | details | | 25-hydroxyvitamin D-1 alpha hydroxylase, mitochondrial | CYP27B1 | 12q13.1-q13.3 | O15528 | details | | Steroid 17-alpha-hydroxylase/17,20 lyase | CYP17A1 | 10q24.3 | P05093 | details | | Sulfite oxidase, mitochondrial | SUOX | 12q13.2 | P51687 | details | | Delta-aminolevulinic acid dehydratase | ALAD | 9q33.1 | P13716 | details | | Guanylate cyclase soluble subunit beta-1 | GUCY1B3 | 4q31.3-q33 | Q02153 | details | | Guanylate cyclase soluble subunit beta-2 | GUCY1B2 | | O75343 | details | | Prostacyclin synthase | PTGIS | 20q13.13 | Q16647 | details | | Cytochrome P450 2E1 | CYP2E1 | 10q24.3-qter | P05181 | details | | Cytochrome P450 4F11 | CYP4F11 | 19p13.1 | Q9HBI6 | details | | Cytochrome P450 3A43 | CYP3A43 | 7q21.1 | Q9HB55 | details | | Cytochrome P450 1B1 | CYP1B1 | 2p22.2 | Q16678 | details | | Cytochrome P450 2D6 | CYP2D6 | 22q13.1 | P10635 | details | | Cytochrome P450 2C18 | CYP2C18 | 10q24 | P33260 | details | | Cytochrome P450 4F12 | CYP4F12 | 19p13.1 | Q9HCS2 | details | | Cytochrome P450 2F1 | CYP2F1 | 19q13.2 | P24903 | details | | Cytochrome P450 4X1 | CYP4X1 | 1 | Q8N118 | details | | Cytochrome P450 2B6 | CYP2B6 | 19q13.2 | P20813 | details | | Cytochrome P450 3A5 | CYP3A5 | 7q21.1 | P20815 | details | | Cytochrome P450 1A1 | CYP1A1 | 15q24.1 | P04798 | details | | Cytochrome P450 2A13 | CYP2A13 | 19q13.2 | Q16696 | details | | Cytochrome P450 3A7 | CYP3A7 | 7q21-q22.1 | P24462 | details | | Cytochrome P450 4B1 | CYP4B1 | 1p34-p12 | P13584 | details | | Cytochrome P450 4Z1 | CYP4Z1 | 1p33 | Q86W10 | details | | Cytochrome P450 1A2 | CYP1A2 | 15q24.1 | P05177 | details | | Cytochrome P450 19A1 | CYP19A1 | 15q21.1 | P11511 | details | | Cytochrome P450 2C8 | CYP2C8 | 10q23.33 | P10632 | details | | Cytochrome P450 2S1 | CYP2S1 | 19q13.1 | Q96SQ9 | details | | Cytochrome P450 4F8 | CYP4F8 | 19p13.1 | P98187 | details | | Cytochrome P450 2J2 | CYP2J2 | 1p31.3-p31.2 | P51589 | details | | Cytochrome P450 2A7 | CYP2A7 | 19q13.2 | P20853 | details | | Cytochrome P450 2A6 | CYP2A6 | 19q13.2 | P11509 | details | | Catalase | CAT | 11p13 | P04040 | details | | Thromboxane-A synthase | TBXAS1 | 7q34-q35 | P24557 | details | | Hemoglobin subunit beta | HBB | 11p15.5 | P68871 | details | | Hemoglobin subunit alpha | HBA1 | 16p13.3 | P69905 | details | | Hemoglobin subunit gamma-1 | HBG1 | 11p15.5 | P69891 | details | | 25-hydroxycholesterol 7-alpha-hydroxylase | CYP7B1 | 8q21.3 | O75881 | details | | Cytochrome c oxidase subunit 1 | MT-CO1 | | P00395 | details | | Cytochrome c | CYCS | 7p15.3 | P99999 | details | | Cytochrome b-245 heavy chain | CYBB | | P04839 | details | | Cytochrome c-type heme lyase | HCCS | Xp22.3 | P53701 | details | | 1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial | CYP24A1 | 20q13 | Q07973 | details | | PR domain zinc finger protein 2 | PRDM2 | 1p36.21 | Q13029 | details | | Cytochrome P450 26A1 | CYP26A1 | 10q23-q24 | O43174 | details | | Serotransferrin | TF | 3q22.1 | P02787 | details | | Myoglobin | MB | 22q13.1 | P02144 | details | | Cytochrome b-245 light chain | CYBA | 16q24 | P13498 | details | | Fatty acid desaturase 1 | FADS1 | 11q12.2-q13.1 | O60427 | details | | Vitamin D 25-hydroxylase | CYP2R1 | 11p15.2 | Q6VVX0 | details | | Hemoglobin subunit gamma-2 | HBG2 | 11p15.5 | P69892 | details | | Cytochrome b5 | CYB5A | 18q23 | P00167 | details | | Fatty acid desaturase 2 | FADS2 | 11q12.2 | O95864 | details | | Cholesterol 24-hydroxylase | CYP46A1 | 14q32.1 | Q9Y6A2 | details | | 24-hydroxycholesterol 7-alpha-hydroxylase | CYP39A1 | 6p21.1-p11.2 | Q9NYL5 | details | | 7-alpha-hydroxycholest-4-en-3-one 12-alpha-hydroxylase | CYP8B1 | 3p22.1 | Q9UNU6 | details | | Cytochrome b5 reductase 4 | CYB5R4 | 6pter-q22.33 | Q7L1T6 | details | | Cytochrome P450 4A22 | CYP4A22 | 1p33 | Q5TCH4 | details | | Indoleamine 2,3-dioxygenase 2 | IDO2 | 8p11.21 | Q6ZQW0 | details | | Cytochrome P450, family 1, subfamily A, polypeptide 1 | CYP1A1 | 15q24.1 | A0N0X8 | details | | NADPH oxidase 5 | NOX5 | 15q23 | Q96PH1 | details | | Peroxidasin homolog | PXDN | 2p25 | Q92626 | details | | Peroxidasin-like protein | PXDNL | 8q11.22-q11.23 | A1KZ92 | details | | Metalloreductase STEAP3 | STEAP3 | 2q14.2 | Q658P3 | details | | NADPH oxidase 1 | NOX1 | | Q9Y5S8 | details | | NADPH oxidase 4 | NOX4 | 11q14.2-q21 | Q9NPH5 | details | | Metalloreductase STEAP1 | STEAP1 | 7q21 | Q9UHE8 | details | | Metalloreductase STEAP2 | STEAP2 | 7q21 | Q8NFT2 | details | | Metalloreductase STEAP4 | STEAP4 | 7q21.12 | Q687X5 | details | | Cytochrome P450 27C1 | CYP27C1 | 2q14.3 | Q4G0S4 | details | | Cytochrome b5 domain-containing protein 1 | CYB5D1 | 17p13.1 | Q6P9G0 | details | | Neuferricin | CYB5D2 | 17p13.2 | Q8WUJ1 | details | | Cytochrome P450 20A1 | CYP20A1 | 2q33.2 | Q6UW02 | details | | Cytochrome P450 26B1 | CYP26B1 | 2p13.2 | Q9NR63 | details | | Cytochrome P450 26C1 | CYP26C1 | 10q23.33 | Q6V0L0 | details | | Cytochrome P450 2U1 | CYP2U1 | 4q25 | Q7Z449 | details | | Cytochrome P450 2W1 | CYP2W1 | 7p22.3 | Q8TAV3 | details | | Cytochrome P450 4F22 | CYP4F22 | | Q6NT55 | details | | Cytochrome P450 4V2 | CYP4V2 | 4q35.2 | Q6ZWL3 | details | | Cytochrome b5 type B | CYB5B | 16q22.1 | O43169 | details | | Cytoglobin | CYGB | 17q25.3 | Q8WWM9 | details | | Fatty acid 2-hydroxylase | FA2H | 16q23 | Q7L5A8 | details | | Fatty acid desaturase 2-like protein | | | A8MWK0 | details | | Fatty acid desaturase 3 | FADS3 | 11q12-q13.1 | Q9Y5Q0 | details | | Hemoglobin subunit theta-1 | HBQ1 | 16p13.3 | P09105 | details | | Hemoglobin subunit zeta | HBZ | 16p13.3 | P02008 | details | | Hemoglobin subunit delta | HBD | 11p15.5 | P02042 | details | | Hemoglobin subunit epsilon | HBE1 | 11p15.5 | P02100 | details | | Hemoglobin subunit mu | HBM | 16p13.3 | Q6B0K9 | details | | E3 ubiquitin-protein ligase HERC2 | HERC2 | 15q13 | O95714 | details | | Neudesin | NENF | 1q32.3 | Q9UMX5 | details | | Neuroglobin | NGB | 14q24.3 | Q9NPG2 | details | | Membrane-associated progesterone receptor component 1 | PGRMC1 | | O00264 | details | | Membrane-associated progesterone receptor component 2 | PGRMC2 | 4q26 | O15173 | details | | Cytochrome P450, family 21, subfamily A, polypeptide 2 | CYP21A2 | 6p21.3 | Q08AG9 | details |
|
|---|
| Receptors | |
| Nitric oxide synthase, brain | NOS1 | 12q24.2-q24.31 | P29475 | details | | Prostaglandin G/H synthase 1 | PTGS1 | 9q32-q33.3 | P23219 | details | | 25-hydroxyvitamin D-1 alpha hydroxylase, mitochondrial | CYP27B1 | 12q13.1-q13.3 | O15528 | details | | Guanylate cyclase soluble subunit beta-1 | GUCY1B3 | 4q31.3-q33 | Q02153 | details | | Prostacyclin synthase | PTGIS | 20q13.13 | Q16647 | details | | 25-hydroxycholesterol 7-alpha-hydroxylase | CYP7B1 | 8q21.3 | O75881 | details | | 1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial | CYP24A1 | 20q13 | Q07973 | details | | Feline leukemia virus subgroup C receptor-related protein 1 | FLVCR1 | 1q32.3 | Q9Y5Y0 | details | | Scavenger receptor cysteine-rich type 1 protein M130 | CD163 | 12p13.3 | Q86VB7 | details | | Peroxidasin homolog | PXDN | 2p25 | Q92626 | details | | Membrane-associated progesterone receptor component 1 | PGRMC1 | | O00264 | details | | Membrane-associated progesterone receptor component 2 | PGRMC2 | 4q26 | O15173 | details |
|
|---|
| Transcriptional Factors | |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0003178 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB031136 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| KEGG Compound ID | C00032 |
|---|
| BioCyc ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Heme |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | Not Available |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17627 |
|---|
| References |
|---|
| General References | Not Available |
|---|

