| Identification |
|---|
| HMDB Protein ID
| CDBP00351 |
| Secondary Accession Numbers
| Not Available |
| Name
| Porphobilinogen deaminase |
| Description
| Not Available |
| Synonyms
|
- HMBS
- Hydroxymethylbilane synthase
- PBG-D
- Pre-uroporphyrinogen synthase
|
| Gene Name
| HMBS |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in hydroxymethylbilane synthase activity |
| Specific Function
| Tetrapolymerization of the monopyrrole PBG into the hydroxymethylbilane pre-uroporphyrinogen in several discrete steps.
|
| GO Classification
|
| Biological Process |
| protoporphyrinogen IX biosynthetic process |
| organ regeneration |
| cellular response to antibiotic |
| cellular response to amine stimulus |
| response to drug |
| astrocyte differentiation |
| cellular response to cytokine stimulus |
| cellular response to lead ion |
| peptidyl-pyrromethane cofactor linkage |
| response to carbohydrate stimulus |
| response to vitamin |
| small molecule metabolic process |
| response to estradiol stimulus |
| response to zinc ion |
| cellular response to dexamethasone stimulus |
| cellular response to arsenic-containing substance |
| response to hypoxia |
| response to amino acid stimulus |
| response to methylmercury |
| response to cobalt ion |
| heme biosynthetic process |
| Cellular Component |
| cytosol |
| condensed chromosome |
| nucleus |
| axon |
| Function |
| catalytic activity |
| transferase activity |
| hydroxymethylbilane synthase activity |
| transferase activity, transferring alkyl or aryl (other than methyl) groups |
| Molecular Function |
| amine binding |
| carboxylic acid binding |
| hydroxymethylbilane synthase activity |
| uroporphyrinogen-III synthase activity |
| coenzyme binding |
| Process |
| protein-cofactor linkage |
| peptidyl-pyrromethane cofactor linkage |
| biosynthetic process |
| cellular biosynthetic process |
| heterocycle biosynthetic process |
| tetrapyrrole biosynthetic process |
| macromolecule modification |
| protein modification process |
| metabolic process |
| macromolecule metabolic process |
| post-translational protein modification |
|
| Cellular Location
|
- Cytoplasm (Probable)
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | protoporphyrin-IX biosynthesis | Not Available | Not Available | | Porphyrin and chlorophyll metabolism | Not Available |  | | Porphyrin Metabolism | 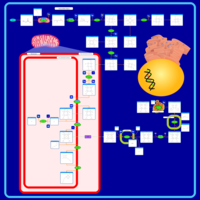 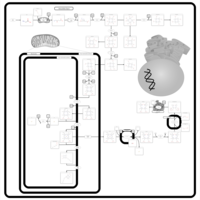 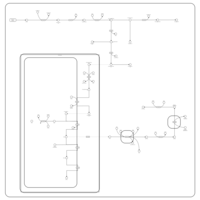 |  | | Acute Intermittent Porphyria | 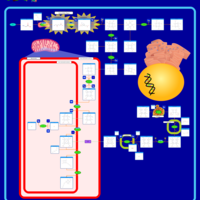   | Not Available | | Porphyria Variegata (PV) |    | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 11 |
| Locus
| 11q23.3 |
| SNPs
| HMBS |
| Gene Sequence
|
>1086 bp
ATGTCTGGTAACGGCAATGCGGCTGCAACGGCGGAAGAAAACAGCCCAAAGATGAGAGTG
ATTCGCGTGGGTACCCGCAAGAGCCAGCTTGCTCGCATACAGACGGACAGTGTGGTGGCA
ACATTGAAAGCCTCGTACCCTGGCCTGCAGTTTGAAATCATTGCTATGTCCACCACAGGG
GACAAGATTCTTGATACTGCACTCTCTAAGATTGGAGAGAAAAGCCTGTTTACCAAGGAG
CTTGAACATGCCCTGGAGAAGAATGAAGTGGACCTGGTTGTTCACTCCTTGAAGGACCTG
CCCACTGTGCTTCCTCCTGGCTTCACCATCGGAGCCATCTGCAAGCGGGAAAACCCTCAT
GATGCTGTTGTCTTTCACCCAAAATTTGTTGGGAAGACCCTAGAAACCCTGCCAGAGAAG
AGTGTGGTGGGAACCAGCTCCCTGCGAAGAGCAGCCCAGCTGCAGAGAAAGTTCCCGCAT
CTGGAGTTCAGGAGTATTCGGGGAAACCTCAACACCCGGCTTCGGAAGCTGGACGAGCAG
CAGGAGTTCAGTGCCATCATCCTGGCAACAGCTGGCCTGCAGCGCATGGGCTGGCACAAC
CGGGTGGGGCAGATCCTGCACCCTGAGGAATGCATGTATGCTGTGGGCCAGGGGGCCTTG
GGCGTGGAAGTGCGAGCCAAGGACCAGGACATCTTGGATCTGGTGGGTGTGCTGCACGAT
CCCGAGACTCTGCTTCGCTGCATCGCTGAAAGGGCCTTCCTGAGGCACCTGGAAGGAGGC
TGCAGTGTGCCAGTAGCCGTGCATACAGCTATGAAGGATGGGCAACTGTACCTGACTGGA
GGAGTCTGGAGTCTAGACGGCTCAGATAGCATACAAGAGACCATGCAGGCTACCATCCAT
GTCCCTGCCCAGCATGAAGATGGCCCTGAGGATGACCCACAGTTGGTAGGCATCACTGCT
CGTAACATTCCACGAGGGCCCCAGTTGGCTGCCCAGAACTTGGGCATCAGCCTGGCCAAC
TTGTTGCTGAGCAAAGGAGCCAAAAACATCCTGGATGTTGCACGGCAGCTTAACGATGCC
CATTAA
|
| Protein Properties |
|---|
| Number of Residues
| 361 |
| Molecular Weight
| 39329.74 |
| Theoretical pI
| 7.174 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Porphobilinogen deaminase
MSGNGNAAATAEENSPKMRVIRVGTRKSQLARIQTDSVVATLKASYPGLQFEIIAMSTTG
DKILDTALSKIGEKSLFTKELEHALEKNEVDLVVHSLKDLPTVLPPGFTIGAICKRENPH
DAVVFHPKFVGKTLETLPEKSVVGTSSLRRAAQLQRKFPHLEFRSIRGNLNTRLRKLDEQ
QEFSAIILATAGLQRMGWHNRVGQILHPEECMYAVGQGALGVEVRAKDQDILDLVGVLHD
PETLLRCIAERAFLRHLEGGCSVPVAVHTAMKDGQLYLTGGVWSLDGSDSIQETMQATIH
VPAQHEDGPEDDPQLVGITARNIPRGPQLAAQNLGISLANLLLSKGAKNILDVARQLNDA
H
|
| External Links |
|---|
| GenBank ID Protein
| 158261573 |
| UniProtKB/Swiss-Prot ID
| P08397 |
| UniProtKB/Swiss-Prot Entry Name
| HEM3_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AK290275 |
| GeneCard ID
| HMBS |
| GenAtlas ID
| HMBS |
| HGNC ID
| HGNC:4982 |
| References |
|---|
| General References
| Not Available |
