| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:45:30 UTC |
|---|
| Updated at | 2020-12-07 19:11:09 UTC |
|---|
| CannabisDB ID | CDB004872 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | S-Adenosylhomocysteine |
|---|
| Description | S-Adenosylhomocysteine, also known as adenosyl-homo-cys or adohcy, belongs to the class of organic compounds known as gamma butyrolactones. Gamma butyrolactones are compounds containing a gamma butyrolactone moiety, which consists of an aliphatic five-member ring with four carbon atoms, one oxygen atom, and bears a ketone group on the carbon adjacent to the oxygen atom. An organic sulfide that is the S-adenosyl derivative of L-homocysteine. S-Adenosylhomocysteine is a very strong basic compound (based on its pKa). S-Adenosylhomocysteine exists in all living species, ranging from bacteria to humans. Outside of the human body, S-Adenosylhomocysteine has been detected, but not quantified in, several different foods, such as arrowhead, guava, purslanes, kiwis, and gooseberries. This could make S-adenosylhomocysteine a potential biomarker for the consumption of these foods. S-Adenosylhomocysteine is a potentially toxic compound. S-Adenosylhomocysteine is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S)-2-Amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)butanoic acid | ChEBI | | 2-S-Adenosyl-L-homocysteine | ChEBI | | Adenosyl-L-homocysteine | ChEBI | | Adenosylhomocysteine | ChEBI | | AdoHcy | ChEBI | | S-(5'-Adenosyl)-L-homocysteine | ChEBI | | S-[1-(Adenin-9-yl)-1,5-dideoxy-beta-D-ribofuranos-5-yl]-L-homocysteine | ChEBI | | SAH | ChEBI | | (2S)-2-Amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)butanoate | Generator | | (2S)-2-Amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulphanyl)butanoate | Generator | | (2S)-2-Amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulphanyl)butanoic acid | Generator | | S-[1-(Adenin-9-yl)-1,5-dideoxy-b-D-ribofuranos-5-yl]-L-homocysteine | Generator | | S-[1-(Adenin-9-yl)-1,5-dideoxy-β-D-ribofuranos-5-yl]-L-homocysteine | Generator | | (S)-5'-(S)-(3-Amino-3-carboxypropyl)-5'-thioadenosine | HMDB | | 5'-Deoxy-S-adenosyl-L-homocysteine | HMDB | | 5'-S-(3-Amino-3-carboxypropyl)-5'-thio-L-adenosine | HMDB | | Adenosyl-homo-cys | HMDB | | Adenosylhomo-cys | HMDB | | Formycinylhomocysteine | HMDB | | L-5'-S-(3-Amino-3-carboxypropyl)-5'-thior-adenosine | HMDB | | L-S-Adenosyl-homocysteine | HMDB | | L-S-Adenosylhomocysteine | HMDB | | S-(5'-Deoxyadenosin-5'-yl)-L-homocysteine | HMDB | | S-(5'-Deoxyadenosine-5')-L-homocysteine | HMDB | | S-Adenosyl-homocysteine | HMDB | | S-Adenosyl-L-homocysteine | HMDB | | Adenosylhomocysteine, S | HMDB | | S Adenosylhomocysteine | HMDB |
|
|---|
| Chemical Formula | C14H20N6O5S |
|---|
| Average Molecular Weight | 384.41 |
|---|
| Monoisotopic Molecular Weight | 384.1216 |
|---|
| IUPAC Name | (2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl}sulfanyl)butanoic acid |
|---|
| Traditional Name | S-adenosyl-L-homocysteine |
|---|
| CAS Registry Number | 979-92-0 |
|---|
| SMILES | N[C@@H](CCSC[C@H]1O[C@H]([C@H](O)[C@@H]1O)N1C=NC2=C1N=CN=C2N)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C14H20N6O5S/c15-6(14(23)24)1-2-26-3-7-9(21)10(22)13(25-7)20-5-19-8-11(16)17-4-18-12(8)20/h4-7,9-10,13,21-22H,1-3,15H2,(H,23,24)(H2,16,17,18)/t6-,7+,9+,10+,13+/m0/s1 |
|---|
| InChI Key | ZJUKTBDSGOFHSH-WFMPWKQPSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as gamma butyrolactones. Gamma butyrolactones are compounds containing a gamma butyrolactone moiety, which consists of an aliphatic five-member ring with four carbon atoms, one oxygen atom, and bears a ketone group on the carbon adjacent to the oxygen atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Lactones |
|---|
| Sub Class | Gamma butyrolactones |
|---|
| Direct Parent | Gamma butyrolactones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Gamma butyrolactone
- Tetrahydrofuran
- Secondary alcohol
- Carboxylic acid ester
- 1,2-diol
- Oxacycle
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Indirect biological role: Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 209 - 211 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | S-Adenosylhomocysteine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0a6u-9853000000-441f171739659ea2b5f6 | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, 3 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-004r-7096060000-d32383fe3b85ef1b9032 | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_1_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_1_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_2_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_2_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_2_6, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_2_7, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_2_8, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_2_9, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_2_10, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_2_11, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_2_12, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_3_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_3_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_3_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | S-Adenosylhomocysteine, TMS_3_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-0319020200-859d659a39a790a582f1 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0a4i-0900000000-53acf4e10681047da062 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-0910000000-5896502e31d66786e12a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-0009000000-01ea51599f8d30dec7fe | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-0409000100-2de58945ab0dfa81f099 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0a4i-1900000000-a640f1a0bafd33afdd2d | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-0910000000-5f685035af9b243ccff2 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-0009000000-8d01fd0969e47d3bcc5c | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-001i-0902000000-c6c0d5529cf011d89f7b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0a4i-0900000000-53acf4e10681047da062 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-001i-0910000000-5896502e31d66786e12a | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-001i-0009000000-01ea51599f8d30dec7fe | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0a4i-1900000000-a640f1a0bafd33afdd2d | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-001i-0910000000-6da47cc70ea7d7eb41f1 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-001i-0009000000-8d01fd0969e47d3bcc5c | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-001i-0902000000-c6c0d5529cf011d89f7b | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-001i-0900000000-978dc4fcc4643522e8ef | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0006-0943000000-80fa5c7d919e1085bc5d | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00di-0900000000-3f143852503952daa0a8 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-03di-0900000000-23694a4b657a3478ad91 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0udr-0980000000-a8f61dcdc8693e2725f5 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00kf-0974000000-b3f3632635b2847bf4d2 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00di-0900000000-188a77c79aaefae8a184 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-0900000000-b848c20d743785117445 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0udr-0980000000-64b7b303fc07300e0782 | 2012-08-31 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, 100%_DMSO, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, 100%_DMSO, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Betaine Metabolism |  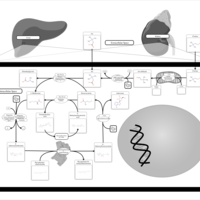  |  | | Methionine Metabolism | 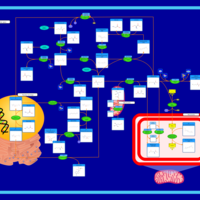 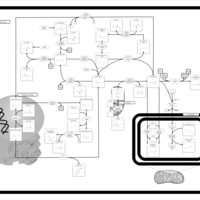 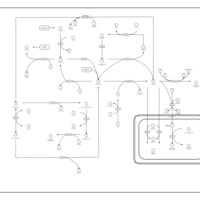 |  | | Carnitine Synthesis | 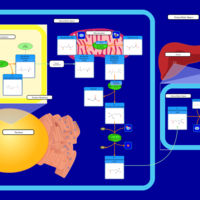 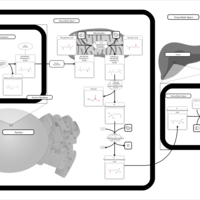 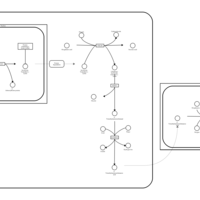 | Not Available | | Glycine and Serine Metabolism | 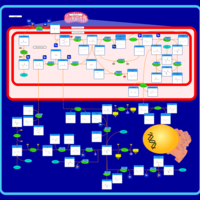 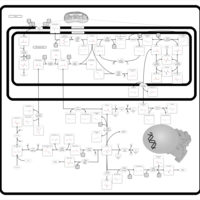 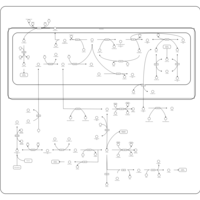 |  | | Arginine and Proline Metabolism | 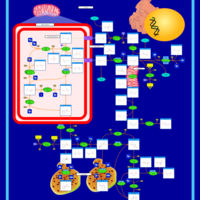 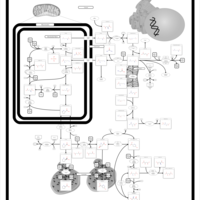  |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Phosphatidylethanolamine N-methyltransferase | PEMT | 17p11.2 | Q9UBM1 | details | | Acetylserotonin O-methyltransferase | ASMT | Xp22.3 or Yp11.3 | P46597 | details | | Catechol O-methyltransferase | COMT | 22q11.21 | P21964 | details | | Histone-lysine N-methyltransferase, H3 lysine-79 specific | DOT1L | 19p13.3 | Q8TEK3 | details | | Glycine N-methyltransferase | GNMT | 6p12 | Q14749 | details | | Histone-lysine N-methyltransferase SETD7 | SETD7 | 4q28 | Q8WTS6 | details | | Phenylethanolamine N-methyltransferase | PNMT | 17q | P11086 | details | | Diphthine synthase | DPH5 | 1p21.2 | Q9H2P9 | details | | Arsenite methyltransferase | AS3MT | 10q24.32 | Q9HBK9 | details | | Mitochondrial tRNA-specific 2-thiouridylase 1 | TRMU | 22q13 | O75648 | details | | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific | NSD1 | 5q35 | Q96L73 | details | | Histone-lysine N-methyltransferase SETDB1 | SETDB1 | 1q21 | Q15047 | details | | Protein-L-isoaspartate(D-aspartate) O-methyltransferase | PCMT1 | 6q24-q25 | P22061 | details | | Protein-S-isoprenylcysteine O-methyltransferase | ICMT | 1p36.21 | O60725 | details | | Indolethylamine N-methyltransferase | INMT | 7p14.3 | O95050 | details | | DNA (cytosine-5)-methyltransferase 3A | DNMT3A | 2p23 | Q9Y6K1 | details | | Hexaprenyldihydroxybenzoate methyltransferase, mitochondrial | COQ3 | 6q16.2 | Q9NZJ6 | details | | Histone-lysine N-methyltransferase EHMT2 | EHMT2 | 6p21.31 | Q96KQ7 | details | | Methionine synthase reductase | MTRR | 5p15.31 | Q9UBK8 | details | | Histone-lysine N-methyltransferase SUV39H1 | SUV39H1 | Xp11.23 | O43463 | details | | Nicotinamide N-methyltransferase | NNMT | 11q23.1 | P40261 | details | | Histone-lysine N-methyltransferase SUV39H2 | SUV39H2 | 10p13 | Q9H5I1 | details | | Histone-lysine N-methyltransferase EHMT1 | EHMT1 | 9q34.3 | Q9H9B1 | details | | Guanidinoacetate N-methyltransferase | GAMT | 19p13.3 | Q14353 | details | | Histamine N-methyltransferase | HNMT | 2q22.1 | P50135 | details | | Thiopurine S-methyltransferase | TPMT | 6p22.3 | P51580 | details | | N6-adenosine-methyltransferase 70 kDa subunit | METTL3 | 14q11.1 | Q86U44 | details | | Putative adenosylhomocysteinase 3 | AHCYL2 | 7q32.1 | Q96HN2 | details | | Adenosylhomocysteinase | AHCY | 20q11.22 | P23526 | details | | Putative adenosylhomocysteinase 2 | AHCYL1 | 1p13.2 | O43865 | details | | Protein arginine N-methyltransferase 5 | PRMT5 | 14q11.2 | O14744 | details | | Histone-lysine N-methyltransferase SETD1A | SETD1A | 16p11.2 | O15047 | details | | Histone-lysine N-methyltransferase MLL | MLL | 11q23 | Q03164 | details | | DNA (cytosine-5)-methyltransferase 3B | DNMT3B | 20q11.2 | Q9UBC3 | details | | tRNA (cytosine(38)-C(5))-methyltransferase | TRDMT1 | 10p15.1 | O14717 | details | | N-lysine methyltransferase SETD8 | SETD8 | 12q24.31 | Q9NQR1 | details | | Histone-lysine N-methyltransferase MLL3 | MLL3 | 7q36.1 | Q8NEZ4 | details | | DNA (cytosine-5)-methyltransferase 1 | DNMT1 | 19p13.2 | P26358 | details | | Histone-lysine N-methyltransferase SETDB2 | SETDB2 | 13q14 | Q96T68 | details | | tRNA (cytosine(34)-C(5))-methyltransferase | NSUN2 | 5p15.31 | Q08J23 | details | | 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial | COQ5 | 12q24.31 | Q5HYK3 | details | | Histone-lysine N-methyltransferase ASH1L | ASH1L | 1q22 | Q9NR48 | details | | Histone-arginine methyltransferase CARM1 | CARM1 | 19p13.2 | Q86X55 | details | | Histone-lysine N-methyltransferase EZH2 | EZH2 | 7q35-q36 | Q15910 | details | | mRNA cap guanine-N7 methyltransferase | RNMT | 18p11.21 | O43148 | details | | Histone-lysine N-methyltransferase MLL2 | MLL2 | 12q13.12 | O14686 | details | | Histone-lysine N-methyltransferase MLL4 | WBP7 | 19q13.1 | Q9UMN6 | details | | Histone-lysine N-methyltransferase MLL5 | MLL5 | 7q22.1 | Q8IZD2 | details | | Histone-lysine N-methyltransferase NSD2 | WHSC1 | 4p16.3 | O96028 | details | | Histone-lysine N-methyltransferase NSD3 | WHSC1L1 | 8p11.2 | Q9BZ95 | details | | Putative tRNA (cytidine(32)/guanosine(34)-2'-O)-methyltransferase | FTSJ1 | Xp11.23 | Q9UET6 | details | | Putative ribosomal RNA methyltransferase 2 | FTSJ2 | 7p22 | Q9UI43 | details | | Histone-lysine N-methyltransferase SETD1B | SETD1B | 12q24.31 | Q9UPS6 | details | | Histone-lysine N-methyltransferase SETD2 | SETD2 | 3p21.31 | Q9BYW2 | details | | Histone-lysine N-methyltransferase SETMAR | SETMAR | 3p26.1 | Q53H47 | details | | Histone-lysine N-methyltransferase SMYD3 | SMYD3 | 1q44 | Q9H7B4 | details | | Histone-lysine N-methyltransferase SUV420H1 | SUV420H1 | 11q13.2 | Q4FZB7 | details | | Histone-lysine N-methyltransferase SUV420H2 | SUV420H2 | 19q13.42 | Q86Y97 | details | | tRNA (guanine(26)-N(2))-dimethyltransferase | TRMT1 | 19p13.2 | Q9NXH9 | details | | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A | TRMT61A | 14q32 | Q96FX7 | details | | tRNA (guanine-N(7)-)-methyltransferase | METTL1 | 12q13 | Q9UBP6 | details | | tRNA (adenine(58)-N(1))-methyltransferase, mitochondrial | TRMT61B | 2p23.2 | Q9BVS5 | details | | tRNA (guanine(37)-N1)-methyltransferase | TRMT5 | 14q23.1 | Q32P41 | details | | Transmembrane O-methyltransferase | LRTOMT | 11q13.4 | Q8WZ04 | details | | DNA (cytosine-5)-methyltransferase 3-like | DNMT3L | | Q9UJW3 | details | | Histone-lysine N-methyltransferase EZH1 | EZH1 | 17q21.1-q21.3 | Q92800 | details | | Leucine carboxyl methyltransferase 1 | LCMT1 | 16p12.1 | Q9UIC8 | details | | tRNA (uracil(54)-C(5))-methyltransferase homolog | TRMT2B | Xq22.1 | Q96GJ1 | details | | Protein arginine N-methyltransferase 1 | PRMT1 | 19q13.3 | Q99873 | details | | Protein arginine N-methyltransferase 2 | PRMT2 | 21q22.3 | P55345 | details | | Protein arginine N-methyltransferase 6 | PRMT6 | 1p13.3 | Q96LA8 | details | | Protein arginine N-methyltransferase 7 | PRMT7 | 16q22.1 | Q9NVM4 | details | | Calmodulin-lysine N-methyltransferase | CAMKMT | 2p21 | Q7Z624 | details | | Probable dimethyladenosine transferase | DIMT1 | 5q12.1 | Q9UNQ2 | details | | HemK methyltransferase family member 1 | HEMK1 | | Q9Y5R4 | details | | Small RNA 2'-O-methyltransferase | HENMT1 | 1p13.3 | Q5T8I9 | details | | Methyltransferase-like protein 2B | METTL2B | | Q6P1Q9 | details | | Methyltransferase-like protein 6 | METTL6 | | Q8TCB7 | details | | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 | FTSJD2 | 6p21.2 | Q8N1G2 | details | | Ribosomal RNA small subunit methyltransferase NEP1 | EMG1 | 12p13.3 | Q92979 | details | | N-terminal Xaa-Pro-Lys N-methyltransferase 1 | NTMT1 | 9q34.11 | Q9BV86 | details | | Alpha N-terminal protein methyltransferase 1B | METTL11B | 1q24.2 | Q5VVY1 | details | | Putative histone-lysine N-methyltransferase PRDM6 | PRDM6 | 5q23.2 | Q9NQX0 | details | | Probable histone-lysine N-methyltransferase PRDM7 | PRDM7 | 16q24.3 | Q9NQW5 | details | | Histone-lysine N-methyltransferase PRDM9 | PRDM9 | 5p14 | Q9NQV7 | details | | Histone-lysine N-methyltransferase setd3 | SETD3 | 14q32.2 | Q86TU7 | details | | N-lysine methyltransferase SMYD2 | SMYD2 | 1q32.3 | Q9NRG4 | details | | Trimethylguanosine synthase | TGS1 | 8q11 | Q96RS0 | details | | Probable tRNA (uracil-O(2)-)-methyltransferase | TRMT44 | 4p16.1 | Q8IYL2 | details | | Uncharacterized methyltransferase WBSCR22 | WBSCR22 | | O43709 | details |
|
|---|
| Transporters | |
|---|
| Metal Bindings | |
| Catechol O-methyltransferase | COMT | 22q11.21 | P21964 | details | | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific | NSD1 | 5q35 | Q96L73 | details | | Histone-lysine N-methyltransferase SETDB1 | SETDB1 | 1q21 | Q15047 | details | | DNA (cytosine-5)-methyltransferase 3A | DNMT3A | 2p23 | Q9Y6K1 | details | | Histone-lysine N-methyltransferase EHMT2 | EHMT2 | 6p21.31 | Q96KQ7 | details | | Methionine synthase reductase | MTRR | 5p15.31 | Q9UBK8 | details | | Histone-lysine N-methyltransferase SUV39H1 | SUV39H1 | Xp11.23 | O43463 | details | | Histone-lysine N-methyltransferase SUV39H2 | SUV39H2 | 10p13 | Q9H5I1 | details | | Histone-lysine N-methyltransferase EHMT1 | EHMT1 | 9q34.3 | Q9H9B1 | details | | PR domain zinc finger protein 2 | PRDM2 | 1p36.21 | Q13029 | details | | Histone-lysine N-methyltransferase MLL | MLL | 11q23 | Q03164 | details | | DNA (cytosine-5)-methyltransferase 3B | DNMT3B | 20q11.2 | Q9UBC3 | details | | Histone-lysine N-methyltransferase MLL3 | MLL3 | 7q36.1 | Q8NEZ4 | details | | DNA (cytosine-5)-methyltransferase 1 | DNMT1 | 19p13.2 | P26358 | details | | Histone-lysine N-methyltransferase SETDB2 | SETDB2 | 13q14 | Q96T68 | details | | Histone-lysine N-methyltransferase ASH1L | ASH1L | 1q22 | Q9NR48 | details | | Histone-lysine N-methyltransferase MLL2 | MLL2 | 12q13.12 | O14686 | details | | Histone-lysine N-methyltransferase MLL4 | WBP7 | 19q13.1 | Q9UMN6 | details | | Histone-lysine N-methyltransferase MLL5 | MLL5 | 7q22.1 | Q8IZD2 | details | | Histone-lysine N-methyltransferase NSD2 | WHSC1 | 4p16.3 | O96028 | details | | Histone-lysine N-methyltransferase NSD3 | WHSC1L1 | 8p11.2 | Q9BZ95 | details | | Histone-lysine N-methyltransferase SETD2 | SETD2 | 3p21.31 | Q9BYW2 | details | | Histone-lysine N-methyltransferase SETMAR | SETMAR | 3p26.1 | Q53H47 | details | | Histone-lysine N-methyltransferase SMYD3 | SMYD3 | 1q44 | Q9H7B4 | details | | tRNA:m(4)X modification enzyme TRM13 homolog | TRMT13 | 1p21.2 | Q9NUP7 | details | | tRNA (guanine(26)-N(2))-dimethyltransferase | TRMT1 | 19p13.2 | Q9NXH9 | details | | DNA (cytosine-5)-methyltransferase 3-like | DNMT3L | | Q9UJW3 | details | | Alkylated DNA repair protein alkB homolog 8 | ALKBH8 | 11q22.3 | Q96BT7 | details | | Small RNA 2'-O-methyltransferase | HENMT1 | 1p13.3 | Q5T8I9 | details | | Putative histone-lysine N-methyltransferase PRDM6 | PRDM6 | 5q23.2 | Q9NQX0 | details | | Histone-lysine N-methyltransferase PRDM9 | PRDM9 | 5p14 | Q9NQV7 | details | | N-lysine methyltransferase SMYD2 | SMYD2 | 1q32.3 | Q9NRG4 | details | | Probable tRNA (uracil-O(2)-)-methyltransferase | TRMT44 | 4p16.1 | Q8IYL2 | details |
|
|---|
| Receptors | |
| Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific | NSD1 | 5q35 | Q96L73 | details | | Histone-arginine methyltransferase CARM1 | CARM1 | 19p13.2 | Q86X55 | details | | Histone-lysine N-methyltransferase EZH2 | EZH2 | 7q35-q36 | Q15910 | details | | Histone-lysine N-methyltransferase MLL2 | MLL2 | 12q13.12 | O14686 | details | | Histone-lysine N-methyltransferase MLL5 | MLL5 | 7q22.1 | Q8IZD2 | details | | Transmembrane O-methyltransferase | LRTOMT | 11q13.4 | Q8WZ04 | details | | Protein arginine N-methyltransferase 1 | PRMT1 | 19q13.3 | Q99873 | details | | Protein arginine N-methyltransferase 2 | PRMT2 | 21q22.3 | P55345 | details |
|
|---|
| Transcriptional Factors | |
| Histone-lysine N-methyltransferase, H3 lysine-79 specific | DOT1L | 19p13.3 | Q8TEK3 | details | | Histone-lysine N-methyltransferase SETD7 | SETD7 | 4q28 | Q8WTS6 | details | | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific | NSD1 | 5q35 | Q96L73 | details | | Histone-lysine N-methyltransferase SETDB1 | SETDB1 | 1q21 | Q15047 | details | | DNA (cytosine-5)-methyltransferase 3A | DNMT3A | 2p23 | Q9Y6K1 | details | | Histone-lysine N-methyltransferase EHMT2 | EHMT2 | 6p21.31 | Q96KQ7 | details | | Histone-lysine N-methyltransferase SUV39H1 | SUV39H1 | Xp11.23 | O43463 | details | | Histone-lysine N-methyltransferase SUV39H2 | SUV39H2 | 10p13 | Q9H5I1 | details | | Histone-lysine N-methyltransferase EHMT1 | EHMT1 | 9q34.3 | Q9H9B1 | details | | Protein arginine N-methyltransferase 5 | PRMT5 | 14q11.2 | O14744 | details | | Histone-lysine N-methyltransferase SETD1A | SETD1A | 16p11.2 | O15047 | details | | PR domain zinc finger protein 2 | PRDM2 | 1p36.21 | Q13029 | details | | Histone-lysine N-methyltransferase MLL | MLL | 11q23 | Q03164 | details | | DNA (cytosine-5)-methyltransferase 3B | DNMT3B | 20q11.2 | Q9UBC3 | details | | N-lysine methyltransferase SETD8 | SETD8 | 12q24.31 | Q9NQR1 | details | | Histone-lysine N-methyltransferase MLL3 | MLL3 | 7q36.1 | Q8NEZ4 | details | | DNA (cytosine-5)-methyltransferase 1 | DNMT1 | 19p13.2 | P26358 | details | | Histone-lysine N-methyltransferase SETDB2 | SETDB2 | 13q14 | Q96T68 | details | | Histone-lysine N-methyltransferase ASH1L | ASH1L | 1q22 | Q9NR48 | details | | Histone-arginine methyltransferase CARM1 | CARM1 | 19p13.2 | Q86X55 | details | | Histone-lysine N-methyltransferase EZH2 | EZH2 | 7q35-q36 | Q15910 | details | | mRNA cap guanine-N7 methyltransferase | RNMT | 18p11.21 | O43148 | details | | Histone-lysine N-methyltransferase MLL2 | MLL2 | 12q13.12 | O14686 | details | | Histone-lysine N-methyltransferase MLL4 | WBP7 | 19q13.1 | Q9UMN6 | details | | Histone-lysine N-methyltransferase MLL5 | MLL5 | 7q22.1 | Q8IZD2 | details | | Histone-lysine N-methyltransferase NSD2 | WHSC1 | 4p16.3 | O96028 | details | | Histone-lysine N-methyltransferase NSD3 | WHSC1L1 | 8p11.2 | Q9BZ95 | details | | Histone-lysine N-methyltransferase SETD1B | SETD1B | 12q24.31 | Q9UPS6 | details | | Histone-lysine N-methyltransferase SETD2 | SETD2 | 3p21.31 | Q9BYW2 | details | | Histone-lysine N-methyltransferase SUV420H1 | SUV420H1 | 11q13.2 | Q4FZB7 | details | | Histone-lysine N-methyltransferase SUV420H2 | SUV420H2 | 19q13.42 | Q86Y97 | details | | DNA (cytosine-5)-methyltransferase 3-like | DNMT3L | | Q9UJW3 | details | | Histone-lysine N-methyltransferase EZH1 | EZH1 | 17q21.1-q21.3 | Q92800 | details | | Protein arginine N-methyltransferase 2 | PRMT2 | 21q22.3 | P55345 | details | | Protein arginine N-methyltransferase 6 | PRMT6 | 1p13.3 | Q96LA8 | details | | Protein arginine N-methyltransferase 7 | PRMT7 | 16q22.1 | Q9NVM4 | details | | Putative histone-lysine N-methyltransferase PRDM6 | PRDM6 | 5q23.2 | Q9NQX0 | details | | Probable histone-lysine N-methyltransferase PRDM7 | PRDM7 | 16q24.3 | Q9NQW5 | details | | Histone-lysine N-methyltransferase PRDM9 | PRDM9 | 5p14 | Q9NQV7 | details | | Histone-lysine N-methyltransferase setd3 | SETD3 | 14q32.2 | Q86TU7 | details | | N-lysine methyltransferase SMYD2 | SMYD2 | 1q32.3 | Q9NRG4 | details | | Trimethylguanosine synthase | TGS1 | 8q11 | Q96RS0 | details |
|
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000939 |
|---|
| DrugBank ID | DB01752 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB031150 |
|---|
| KNApSAcK ID | C00007230 |
|---|
| Chemspider ID | 388301 |
|---|
| KEGG Compound ID | C00021 |
|---|
| BioCyc ID | ADENOSYL-HOMO-CYS |
|---|
| BiGG ID | 33543 |
|---|
| Wikipedia Link | S-Adenosyl-L-homocysteine |
|---|
| METLIN ID | 296 |
|---|
| PubChem Compound | 439155 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16680 |
|---|
| References |
|---|
| General References | Not Available |
|---|


