| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:08:19 UTC |
|---|
| Updated at | 2020-11-18 16:39:19 UTC |
|---|
| CannabisDB ID | CDB005090 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Potassium |
|---|
| Description | Potassium, also known as K+ or potassium ion, belongs to the class of inorganic compounds known as homogeneous alkali metal compounds. These are inorganic compounds containing only metal atoms,with the largest atom being a alkali metal atom. Potassium is a weakly acidic compound (based on its pKa). Potassium exists in all living species, ranging from bacteria to humans. In humans, potassium is involved in the metabolic disorder called the phenytoin (antiarrhythmic) action pathway. Outside of the human body, Potassium is found, on average, in the highest concentration within a few different foods, such as kombus, milk (cow), and garden tomato (var.) and in a lower concentration in yogurts, strawberries, and zwiebacks. Potassium has also been detected, but not quantified in, several different foods, such as sourdocks, half-highbush blueberries, wax apples, jostaberries, and medlars. This could make potassium a potential biomarker for the consumption of these foods. Potassium is a potentially toxic compound. Potassium is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| K(+) | ChEBI | | K+ | ChEBI | | POTASSIUM ion | ChEBI | | Potassium(1+) | Kegg | | Nabumeton a | Kegg | | Kalium | HMDB | | Potassium (ion) | HMDB | | Potassium (k+) | HMDB | | Potassium cation | HMDB | | Potassium ion (k+) | HMDB | | Potassium ion (K1+) | HMDB | | Potassium ion(+) | HMDB | | Potassium ion(1+) | HMDB | | Potassium monocation | HMDB | | Potassium(+) | HMDB | | Potassium(1+) ion | HMDB | | Potassium(I) cation | HMDB | | Liver regeneration factor 1 | HMDB | | LRF-1 | HMDB | | LRF1 Transcription factor | HMDB |
|
|---|
| Chemical Formula | K |
|---|
| Average Molecular Weight | 39.1 |
|---|
| Monoisotopic Molecular Weight | 38.9637 |
|---|
| IUPAC Name | potassium(1+) ion |
|---|
| Traditional Name | potassium(1+) ion |
|---|
| CAS Registry Number | 7440-09-7 |
|---|
| SMILES | [K+] |
|---|
| InChI Identifier | InChI=1S/K/q+1 |
|---|
| InChI Key | NPYPAHLBTDXSSS-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of inorganic compounds known as homogeneous alkali metal compounds. These are inorganic compounds containing only metal atoms,with the largest atom being a alkali metal atom. |
|---|
| Kingdom | Inorganic compounds |
|---|
| Super Class | Homogeneous metal compounds |
|---|
| Class | Homogeneous alkali metal compounds |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Homogeneous alkali metal compounds |
|---|
| Alternative Parents | Not Available |
|---|
| Substituents | |
|---|
| Molecular Framework | Not Available |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 63.2 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | Not Available |
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-9000000000-560fc6f738e9570ec8a2 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-9000000000-560fc6f738e9570ec8a2 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-9000000000-560fc6f738e9570ec8a2 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-9000000000-bcbe5ea7d5d32f6a9598 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9000000000-bcbe5ea7d5d32f6a9598 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000i-9000000000-bcbe5ea7d5d32f6a9598 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-9000000000-e11fed6c31524d9e4d2c | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-9000000000-e11fed6c31524d9e4d2c | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-9000000000-e11fed6c31524d9e4d2c | 2021-09-22 | View Spectrum |
|
|---|
| NMR | Not Available |
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Lactose Degradation | 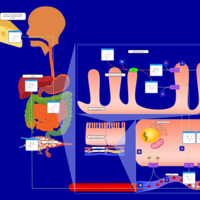 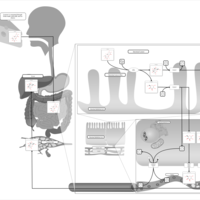  | Not Available | | Trehalose Degradation | 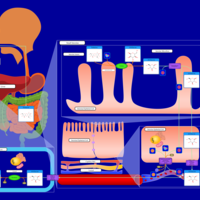 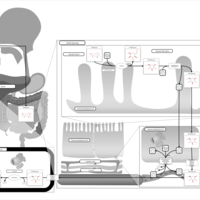 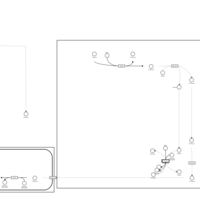 | Not Available | | Spermidine and Spermine Biosynthesis | 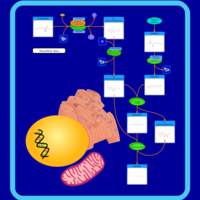 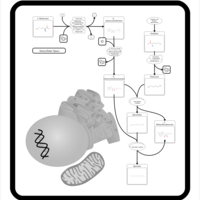 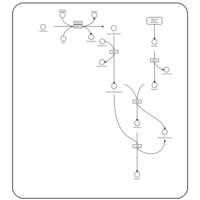 | Not Available | | Pyruvate Metabolism |  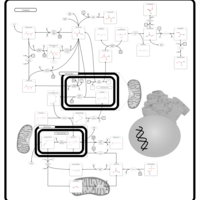  |  | | Leigh Syndrome | 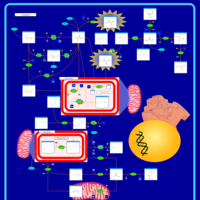 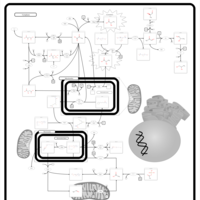 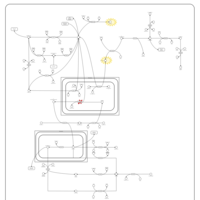 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial | BCKDHA | 19q13.1-q13.2 | P12694 | details | | S-adenosylmethionine synthase isoform type-2 | MAT2A | 2p11.2 | P31153 | details | | S-adenosylmethionine synthase isoform type-1 | MAT1A | 10q22 | Q00266 | details | | Pyruvate kinase isozymes M1/M2 | PKM | 15q22 | P14618 | details | | Pyruvate kinase isozymes R/L | PKLR | 1q21 | P30613 | details | | Inosine-5'-monophosphate dehydrogenase 2 | IMPDH2 | 3p21.2 | P12268 | details | | Inosine-5'-monophosphate dehydrogenase 1 | IMPDH1 | 7q31.3-q32 | P20839 | details | | Pyridoxal kinase | PDXK | 21q22.3 | O00764 | details | | Sodium/potassium-transporting ATPase subunit beta-3 | ATP1B3 | 3q23 | P54709 | details | | Sodium/potassium-transporting ATPase subunit beta-2 | ATP1B2 | 17p13.1 | P14415 | details | | Sodium/potassium-transporting ATPase subunit beta-1 | ATP1B1 | 1q24 | P05026 | details | | Potassium-transporting ATPase alpha chain 2 | ATP12A | 13q12.1-q12.3 | P54707 | details | | Sodium/potassium-transporting ATPase subunit alpha-1 | ATP1A1 | 1p21 | P05023 | details | | Sodium/potassium-transporting ATPase subunit alpha-3 | ATP1A3 | 19q13.31 | P13637 | details | | Potassium-transporting ATPase subunit beta | ATP4B | 13q34 | P51164 | details | | Potassium-transporting ATPase alpha chain 1 | ATP4A | 19q13.1 | P20648 | details | | Sodium/potassium-transporting ATPase subunit alpha-2 | ATP1A2 | 1q23.2 | P50993 | details | | Sodium/potassium-transporting ATPase subunit alpha-4 | ATP1A4 | 1q23.2 | Q13733 | details | | NADPH-dependent diflavin oxidoreductase 1 | NDOR1 | 9q34.3 | Q9UHB4 | details | | Acyl-coenzyme A synthetase ACSM2B, mitochondrial | ACSM2B | 16p12.3 | Q68CK6 | details | | Tubulin--tyrosine ligase | TTL | 2q13 | Q8NG68 | details | | Pyruvate kinase | | | Q16715 | details |
|
|---|
| Transporters | |
| Sodium/potassium-transporting ATPase subunit beta-3 | ATP1B3 | 3q23 | P54709 | details | | Sodium/potassium-transporting ATPase subunit beta-2 | ATP1B2 | 17p13.1 | P14415 | details | | Sodium/potassium-transporting ATPase subunit beta-1 | ATP1B1 | 1q24 | P05026 | details | | Potassium-transporting ATPase alpha chain 2 | ATP12A | 13q12.1-q12.3 | P54707 | details | | Protein ATP1B4 | ATP1B4 | | Q9UN42 | details | | Sodium/potassium-transporting ATPase subunit alpha-1 | ATP1A1 | 1p21 | P05023 | details | | Sodium/potassium-transporting ATPase subunit alpha-3 | ATP1A3 | 19q13.31 | P13637 | details | | Potassium-transporting ATPase subunit beta | ATP4B | 13q34 | P51164 | details | | Potassium-transporting ATPase alpha chain 1 | ATP4A | 19q13.1 | P20648 | details | | Sodium/potassium-transporting ATPase subunit alpha-2 | ATP1A2 | 1q23.2 | P50993 | details | | Calcium-activated potassium channel subunit alpha-1 | KCNMA1 | 10q22.3 | Q12791 | details | | Potassium voltage-gated channel subfamily H member 2 | KCNH2 | 7q36.1 | Q12809 | details | | Inward rectifier potassium channel 2 | KCNJ2 | 17q24.3 | P63252 | details | | Potassium voltage-gated channel subfamily KQT member 1 | KCNQ1 | 11p15.5 | P51787 | details | | ATP-sensitive inward rectifier potassium channel 1 | KCNJ1 | 11q24 | P48048 | details | | ATP-sensitive inward rectifier potassium channel 11 | KCNJ11 | 11p15.1 | Q14654 | details | | Sodium/potassium-transporting ATPase subunit alpha-4 | ATP1A4 | 1q23.2 | Q13733 | details | | Calcium-activated potassium channel subunit beta-3 | KCNMB3 | 3q26.3-q27 | Q9NPA1 | details | | Potassium voltage-gated channel subfamily E member 3 | KCNE3 | 11q13.4 | Q9Y6H6 | details | | Potassium voltage-gated channel subfamily H member 1 | KCNH1 | 1q32.2 | O95259 | details | | Potassium voltage-gated channel subfamily E member 1 | KCNE1 | 21q22.1-q22.2|21q22.12 | P15382 | details | | Potassium voltage-gated channel subfamily KQT member 2 | KCNQ2 | 20q13.3 | O43526 | details | | Potassium voltage-gated channel subfamily A member 1 | KCNA1 | 12p13.32 | Q09470 | details | | Potassium voltage-gated channel subfamily KQT member 3 | KCNQ3 | 8q24 | O43525 | details | | Potassium voltage-gated channel subfamily E member 2 | KCNE2 | 21q22.12 | Q9Y6J6 | details | | Potassium voltage-gated channel subfamily D member 2 | KCND2 | 7q31 | Q9NZV8 | details | | Potassium voltage-gated channel subfamily D member 3 | KCND3 | 1p13.3 | Q9UK17 | details | | Potassium voltage-gated channel subfamily KQT member 5 | KCNQ5 | 6q14 | Q9NR82 | details | | Potassium voltage-gated channel subfamily A member 4 | KCNA4 | 11p14 | P22459 | details | | Potassium voltage-gated channel subfamily A member 3 | KCNA3 | 1p13.3 | P22001 | details | | Potassium voltage-gated channel subfamily C member 3 | KCNC3 | 19q13.33 | Q14003 | details | | Potassium voltage-gated channel subfamily KQT member 4 | KCNQ4 | 1p34 | P56696 | details | | Potassium voltage-gated channel subfamily C member 4 | KCNC4 | 1p21 | Q03721 | details | | Potassium channel subfamily K member 1 | KCNK1 | 1q42-q43 | O00180 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | |
| Sodium/potassium-transporting ATPase subunit alpha-3 | ATP1A3 | 19q13.31 | P13637 | details | | Gamma-aminobutyric acid type B receptor subunit 1 | GABBR1 | 6p21.31 | Q9UBS5 | details | | Steroidogenic factor 1 | NR5A1 | 9q33 | Q13285 | details |
|
|---|
| Transcriptional Factors | |
| Potassium voltage-gated channel subfamily H member 2 | KCNH2 | 7q36.1 | Q12809 | details | | Steroidogenic factor 1 | NR5A1 | 9q33 | Q13285 | details | | Potassium voltage-gated channel subfamily H member 1 | KCNH1 | 1q32.2 | O95259 | details |
|
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 46.935 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 44.449 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 41.741 mg/g dry wt | | details | | Quadra | Detected and Quantified | 36.899 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 44.703 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 29.918 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000586 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB003521 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| KEGG Compound ID | C00238 |
|---|
| BioCyc ID | Not Available |
|---|
| BiGG ID | 34349 |
|---|
| Wikipedia Link | Potassium |
|---|
| METLIN ID | 3197 |
|---|
| PubChem Compound | 813 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 29103 |
|---|
| References |
|---|
| General References | Not Available |
|---|
