| Identification |
|---|
| HMDB Protein ID
| CDBP00710 |
| Secondary Accession Numbers
| Not Available |
| Name
| Pyruvate kinase isozymes M1/M2 |
| Description
| Not Available |
| Synonyms
|
- CTHBP
- Cytosolic thyroid hormone-binding protein
- OIP-3
- Opa-interacting protein 3
- Pyruvate kinase 2/3
- Pyruvate kinase muscle isozyme
- THBP1
- Thyroid hormone-binding protein 1
- Tumor M2-PK
- p58
|
| Gene Name
| PKM |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in magnesium ion binding |
| Specific Function
| Glycolytic enzyme that catalyzes the transfer of a phosphoryl group from phosphoenolpyruvate (PEP) to ADP, generating ATP. Stimulates POU5F1-mediated transcriptional activation. Plays a general role in caspase independent cell death of tumor cells. The ratio betwween the highly active tetrameric form and nearly inactive dimeric form determines whether glucose carbons are channeled to biosynthetic processes or used for glycolytic ATP production. The transition between the 2 forms contributes to the control of glycolysis and is important for tumor cell proliferation and survival.
|
| GO Classification
|
| Biological Process |
| glycolysis |
| response to insulin stimulus |
| liver development |
| response to nutrient |
| response to hypoxia |
| pyruvate biosynthetic process |
| small molecule metabolic process |
| ATP biosynthetic process |
| programmed cell death |
| response to gravity |
| response to muscle inactivity |
| skeletal muscle tissue regeneration |
| organ regeneration |
| Cellular Component |
| cytosol |
| mitochondrion |
| extracellular vesicular exosome |
| plasma membrane |
| nucleus |
| flagellum |
| Function |
| catalytic activity |
| transferase activity |
| transferase activity, transferring phosphorus-containing groups |
| kinase activity |
| magnesium ion binding |
| alkali metal ion binding |
| potassium ion binding |
| pyruvate kinase activity |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| Molecular Function |
| potassium ion binding |
| pyruvate kinase activity |
| magnesium ion binding |
| ATP binding |
| ADP binding |
| Process |
| monosaccharide metabolic process |
| hexose metabolic process |
| glucose metabolic process |
| glucose catabolic process |
| glycolysis |
| metabolic process |
| small molecule metabolic process |
| alcohol metabolic process |
|
| Cellular Location
|
- Nucleus
- Cytoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Glycolysis | 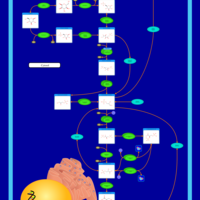 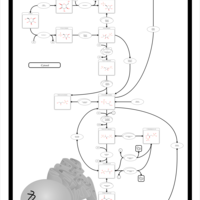 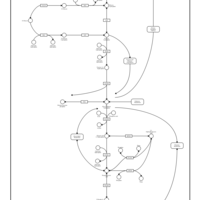 |  | | Glycolysis / Gluconeogenesis | Not Available |  | | Type II diabetes mellitus | Not Available | 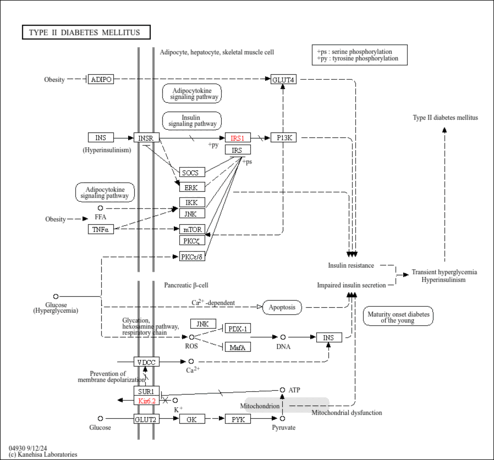 | | Viral carcinogenesis | Not Available |  | | Warburg Effect | 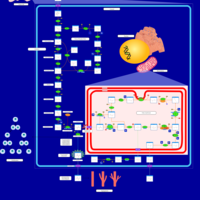 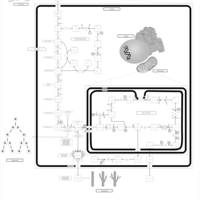  | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 15 |
| Locus
| 15q22 |
| SNPs
| PKM2 |
| Gene Sequence
|
>1596 bp
ATGTCGAAGCCCCATAGTGAAGCCGGGACTGCCTTCATTCAGACCCAGCAGCTGCACGCA
GCCATGGCTGACACATTCCTGGAGCACATGTGCCGCCTGGACATTGATTCACCACCCATC
ACAGCCCGGAACACTGGCATCATCTGTACCATTGGCCCAGCTTCCCGATCAGTGGAGACG
TTGAAGGAGATGATTAAGTCTGGAATGAATGTGGCTCGTCTGAACTTCTCTCATGGAACT
CATGAGTACCATGCGGAGACCATCAAGAATGTGCGCACAGCCACGGAAAGCTTTGCTTCT
GACCCCATCCTCTACCGGCCCGTTGCTGTGGCTCTAGACACTAAAGGACCTGAGATCCGA
ACTGGGCTCATCAAGGGCAGCGGCACTGCAGAGGTGGAGCTGAAGAAGGGAGCCACTCTC
AAAATCACGCTGGATAACGCCTACATGGAAAAGTGTGACGAGAACATCCTGTGGCTGGAC
TACAAGAACATCTGCAAGGTGGTGGAAGTGGGCAGCAAGATCTACGTGGATGATGGGCTT
ATTTCTCTCCAGGTGAAGCAGAAAGGTGCCGACTTCCTGGTGACGGAGGTGGAAAATGGT
GGCTCCTTGGGCAGCAAGAAGGGTGTGAACCTTCCTGGGGCTGCTGTGGACTTGCCTGCT
GTGTCGGAGAAGGACATCCAGGATCTGAAGTTTGGGGTCGAGCAGGATGTTGATATGGTG
TTTGCGTCATTCATCCGCAAGGCATCTGATGTCCATGAAGTTAGGAAGGTCCTGGGAGAG
AAGGGAAAGAACATCAAGATTATCAGCAAAATCGAGAATCATGAGGGGGTTCGGAGGTTT
GATGAAATCCTGGAGGCCAGTGATGGGATCATGGTGGCTCGTGGTGATCTAGGCATTGAG
ATTCCTGCAGAGAAGGTCTTCCTTGCTCAGAAGATGATGATTGGACGGTGCAACCGAGCT
GGGAAGCCTGTCATCTGTGCTACTCAGATGCTGGAGAGCATGATCAAGAAGCCCCGCCCC
ACTCGGGCTGAAGGCAGTGATGTGGCCAATGCAGTCCTGGATGGAGCCGACTGCATCATG
CTGTCTGGAGAAACAGCCAAAGGGGACTATCCTCTGGAGGCTGTGCGCATGCAGCACCTG
ATTGCCCGTGAGGCAGAGGCTGCCATCTACCACTTGCAATTATTTGAGGAACTCCGCCGC
CTGGCGCCCATTACCAGCGACCCCACAGAAGCCACCGCCGTGGGTGCCGTGGAGGCCTCC
TTCAAGTGCTGCAGTGGGGCCATAATCGTCCTCACCAAGTCTGGCAGGTCTGCTCACCAG
GTGGCCAGATACCGCCCACGTGCCCCCATCATTGCTGTGACCCGGAATCCCCAGACAGCT
CGTCAGGCCCACCTGTACCGTGGCATCTTCCCTGTGCTGTGCAAGGACCCAGTCCAGGAG
GCCTGGGCTGAGGACGTGGACCTCCGGGTGAACTTTGCCATGAATGTTGGCAAGGCCCGA
GGCTTCTTCAAGAAGGGAGATGTGGTCATTGTGCTGACCGGATGGCGCCCTGGCTCCGGC
TTCACCAACACCATGCGTGTTGTTCCTGTGCCGTGA
|
| Protein Properties |
|---|
| Number of Residues
| 531 |
| Molecular Weight
| 65930.14 |
| Theoretical pI
| 7.87 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Pyruvate kinase isozymes M1/M2
MSKPHSEAGTAFIQTQQLHAAMADTFLEHMCRLDIDSPPITARNTGIICTIGPASRSVET
LKEMIKSGMNVARLNFSHGTHEYHAETIKNVRTATESFASDPILYRPVAVALDTKGPEIR
TGLIKGSGTAEVELKKGATLKITLDNAYMEKCDENILWLDYKNICKVVEVGSKIYVDDGL
ISLQVKQKGADFLVTEVENGGSLGSKKGVNLPGAAVDLPAVSEKDIQDLKFGVEQDVDMV
FASFIRKASDVHEVRKVLGEKGKNIKIISKIENHEGVRRFDEILEASDGIMVARGDLGIE
IPAEKVFLAQKMMIGRCNRAGKPVICATQMLESMIKKPRPTRAEGSDVANAVLDGADCIM
LSGETAKGDYPLEAVRMQHLIAREAEAAIYHLQLFEELRRLAPITSDPTEATAVGAVEAS
FKCCSGAIIVLTKSGRSAHQVARYRPRAPIIAVTRNPQTARQAHLYRGIFPVLCKDPVQE
AWAEDVDLRVNFAMNVGKARGFFKKGDVVIVLTGWRPGSGFTNTMRVVPVP
|
| External Links |
|---|
| GenBank ID Protein
| 33286418 |
| UniProtKB/Swiss-Prot ID
| P14618 |
| UniProtKB/Swiss-Prot Entry Name
| KPYM_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| NM_002654.3 |
| GeneCard ID
| PKM2 |
| GenAtlas ID
| PKM2 |
| HGNC ID
| HGNC:9021 |
| References |
|---|
| General References
| Not Available |


