| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:53:25 UTC |
|---|
| Updated at | 2020-12-07 19:11:21 UTC |
|---|
| CannabisDB ID | CDB004947 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Hydrogen peroxide |
|---|
| Description | Hydrogen peroxide, also known as oxydol or dihydrogen dioxide, belongs to the class of inorganic compounds known as homogeneous other non-metal compounds. These are inorganic non-metallic compounds in which the largest atom belongs to the class of 'other non-metals'. Hydrogen peroxide is an extremely weak basic (essentially neutral) compound (based on its pKa). Hydrogen peroxide exists in all living species, ranging from bacteria to humans. In humans, hydrogen peroxide is involved in disulfiram action pathway. Outside of the human body, Hydrogen peroxide has been detected, but not quantified in, several different foods, such as tree ferns, napa cabbages, lingonberries, chives, and sesames. This could make hydrogen peroxide a potential biomarker for the consumption of these foods. An inorganic peroxide consisting of two hydroxy groups joined by a covalent oxygen-oxygen single bond. Hydrogen peroxide is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| [OH(OH)] | ChEBI | | Dihydrogen dioxide | ChEBI | | H2O2 | ChEBI | | HOOH | ChEBI | | Oxydol | ChEBI | | Perhydrol | ChEBI | | Oxyfull | Kegg | | Adeka super el | HMDB | | Albone | HMDB | | Albone 35 | HMDB | | Albone DS | HMDB | | Anti-keim 50 | HMDB | | Asepticper | HMDB | | Baquashock | HMDB | | CIX | HMDB | | Clarigel gold | HMDB | | Crestal whitestrips | HMDB | | Crystacide | HMDB | | Dentasept | HMDB | | Deslime LP | HMDB | | Hioxyl | HMDB | | Hipox | HMDB | | Hybrite | HMDB | | Hydrogen dioxide | HMDB | | Inhibine | HMDB | | Lase peroxide | HMDB | | Lensan a | HMDB | | Magic bleaching | HMDB | | Metrokur | HMDB | | Mirasept | HMDB | | Nite white excel 2 | HMDB | | Odosat D | HMDB | | Opalescence xtra | HMDB | | Oxigenal | HMDB | | Oxysept | HMDB | | Oxysept I | HMDB | | Pegasyl | HMDB | | Perone | HMDB | | Peroxaan | HMDB | | Peroxclean | HMDB | | Quasar brite | HMDB | | Select bleach | HMDB | | Superoxol | HMDB | | T-Stuff | HMDB | | Whiteness HP | HMDB | | Whitespeed | HMDB | | Xtra white | HMDB | | Hydrogen peroxide (H2O2) | HMDB | | Hydroperoxide | HMDB | | Peroxide, hydrogen | HMDB |
|
|---|
| Chemical Formula | H2O2 |
|---|
| Average Molecular Weight | 34.01 |
|---|
| Monoisotopic Molecular Weight | 34.0055 |
|---|
| IUPAC Name | peroxol |
|---|
| Traditional Name | hydrogen peroxide |
|---|
| CAS Registry Number | 7722-84-1 |
|---|
| SMILES | OO |
|---|
| InChI Identifier | InChI=1S/H2O2/c1-2/h1-2H |
|---|
| InChI Key | MHAJPDPJQMAIIY-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of inorganic compounds known as homogeneous other non-metal compounds. These are inorganic non-metallic compounds in which the largest atom belongs to the class of 'other non-metals'. |
|---|
| Kingdom | Inorganic compounds |
|---|
| Super Class | Homogeneous non-metal compounds |
|---|
| Class | Homogeneous other non-metal compounds |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Homogeneous other non-metal compounds |
|---|
| Alternative Parents | Not Available |
|---|
| Substituents | - Homogeneous other non metal
|
|---|
| Molecular Framework | Not Available |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Biological location: Source: |
|---|
| Role | Industrial application: |
|---|
| Physical Properties |
|---|
| State | Liquid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | -0.43 °C | Not Available | | Boiling Point | 150.2 °C | Wikipedia | | Water Solubility | 1000 mg/mL at 25 °C | Not Available | | logP | -0.43 | Wikipedia |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Hydrogen peroxide, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-001i-9000000000-4749d46780bc552fb9af | Spectrum | | Predicted GC-MS | Hydrogen peroxide, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Hydrogen peroxide, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-9000000000-50a90bb1548c24e83fbe | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-9000000000-50a90bb1548c24e83fbe | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-9000000000-50a90bb1548c24e83fbe | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-9000000000-a9a93dd42f2cfa0b34c4 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-9000000000-a9a93dd42f2cfa0b34c4 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-9000000000-a9a93dd42f2cfa0b34c4 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-9000000000-d77c0031598ec85f7286 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-9000000000-d77c0031598ec85f7286 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-9000000000-d77c0031598ec85f7286 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-9000000000-86b45185e1deeaca15e1 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-9000000000-86b45185e1deeaca15e1 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-9000000000-86b45185e1deeaca15e1 | 2021-09-22 | View Spectrum |
|
|---|
| NMR | Not Available |
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Phenylalanine and Tyrosine Metabolism | 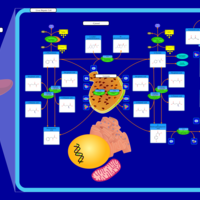 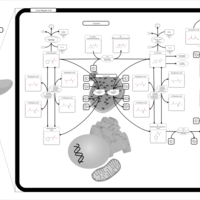 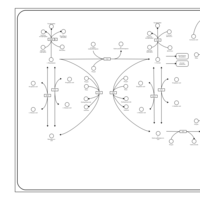 |  | | Histidine Metabolism | 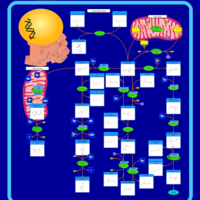 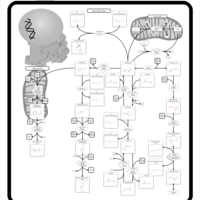 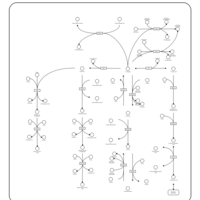 |  | | D-Arginine and D-Ornithine Metabolism |    | 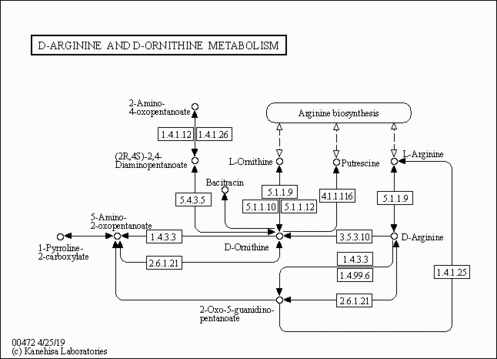 | | Riboflavin Metabolism |   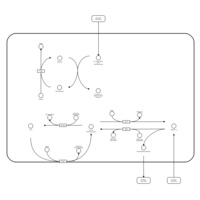 |  | | Ethanol Degradation | 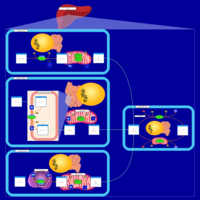 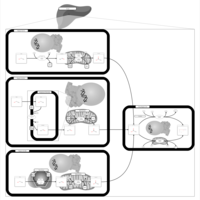 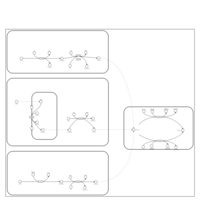 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0003125 |
|---|
| DrugBank ID | DB11091 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB014562 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 763 |
|---|
| KEGG Compound ID | C00027 |
|---|
| BioCyc ID | HYDROGEN-PEROXIDE |
|---|
| BiGG ID | 33570 |
|---|
| Wikipedia Link | Hydrogen_peroxide |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 784 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16240 |
|---|
| References |
|---|
| General References | Not Available |
|---|



