| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:21:31 UTC |
|---|
| Updated at | 2020-12-07 19:06:54 UTC |
|---|
| CannabisDB ID | CDB006141 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Ethanol |
|---|
| Description | Ethanol, also known as ethyl alcohol or alcohol, belongs to the class of organic compounds known as primary alcohols. Primary alcohols are compounds having a primary alcohol functional group, with the general structure RCOH (R=alkyl, aryl). Ethanol is a clear, colorless liquid rapidly absorbed from the gastrointestinal tract and distributed throughout the body. It has bactericidal activity and is used often as a topical disinfectant. It is widely used as a solvent and preservative in pharmaceutical preparations as well as serving as the primary ingredient in alcoholic beverages. Indeed, ethanol has widespread use as a solvent of substances intended for human contact or consumption, including scents, flavorings, colorings, and medicines. Ethanol has a depressive effect on the central nervous system and because of its psychoactive effects, it is considered a drug. Ethanol has a complex mode of action and affects multiple systems in the brain, most notably it acts as an agonist to the GABA receptors. Death from ethanol consumption is possible when blood alcohol level reaches 0.4%. A blood level of 0.5% or more is commonly fatal. Levels of even less than 0.1% can cause intoxication, with unconsciousness often occurring at 0.3-0.4 %. Ethanol is metabolized by the body as an energy-providing carbohydrate nutrient, as it metabolizes into acetyl CoA, an intermediate common with glucose metabolism, that can be used for energy in the citric acid cycle or for biosynthesis. Ethanol within the human body is converted into acetaldehyde by alcohol dehydrogenase and then into acetic acid by acetaldehyde dehydrogenase. The product of the first step of this breakdown, acetaldehyde, is more toxic than ethanol. Acetaldehyde is linked to most of the adverse clinical effects of alcohol. Ethanol has been shown to increase the risk of developing cirrhosis of the liver, multiple forms of cancer, and alcoholism. Industrially, ethanol is produced both as a petrochemical, through the hydration of ethylene, and biologically, by fermenting sugars with yeast. Small amounts of ethanol are endogenously produced by gut microflora through anaerobic fermentation. However, most ethanol detected in biofluids and tissues likely comes from consumption of alcoholic beverages. Absolute ethanol or anhydrous alcohol generally refers to purified ethanol, containing no more than one percent water. Absolute alcohol is not intended for human consumption. It often contains trace amounts of toxic benzene (used to remove water by azeotropic distillation). Consumption of this form of ethanol can be fatal over a short time period. Generally absolute or pure ethanol is used as a solvent for lab and industrial settings where water will disrupt a desired reaction. Pure ethanol is classed as 200 proof in the USA and Canada, equivalent to 175 degrees proof in the UK system. Ethanol is a general biomarker for the consumption of alcohol. Ethanol is also a metabolite of Hansenula and Saccharomyces (PMID: 14613880 ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-Hydroxyethane | ChEBI | | [CH2Me(OH)] | ChEBI | | [OEtH] | ChEBI | | Alcohol | ChEBI | | Alcohol etilico | ChEBI | | Alcool ethylique | ChEBI | | Alkohol | ChEBI | | Aethanol | ChEBI | | Aethylalkohol | ChEBI | | C2H5OH | ChEBI | | Dehydrated ethanol | ChEBI | | Etanol | ChEBI | | Ethyl alcohol | ChEBI | | EtOH | ChEBI | | Hydroxyethane | ChEBI | | Methylcarbinol | ChEBI | | Spiritus vini | ChEBI | | Anhydrous ethanol | Kegg | | Absolute alcohol | MeSH | | Alcohol, absolute | MeSH | | Alcohol, grain | MeSH | | Grain alcohol | MeSH | | Alcohol, ethyl | MeSH | | Absolute ethanol | HMDB | | Absolute ethyl alcohol | HMDB | | Alcare hand degermer | HMDB | | Alcohols | HMDB | | Alcool etilico | HMDB | | Algrain | HMDB | | Alkoholu etylowego | HMDB | | Anhydrol | HMDB | | Anhydrous alcohol | HMDB | | Cologne spirit | HMDB | | Cologne spirits | HMDB | | Dehydrated alcohol | HMDB | | Denatured alcohol | HMDB | | Denatured ethanol | HMDB | | Desinfektol el | HMDB | | Diluted alcohol | HMDB | | Distilled spirits | HMDB | | Ethanol 200 proof | HMDB | | Ethanol solution | HMDB | | Ethicap | HMDB | | Ethyl alc | HMDB | | Ethyl alcohol anhydrous | HMDB | | Ethyl alcohol in alcoholic beverages | HMDB | | Ethyl alcohol usp | HMDB | | Ethyl hydrate | HMDB | | Ethyl hydroxide | HMDB | | Fermentation alcohol | HMDB | | Hinetoless | HMDB | | Infinity pure | HMDB | | Jaysol | HMDB | | Jaysol S | HMDB | | Lux | HMDB | | Molasses alcohol | HMDB | | potato Alcohol | HMDB | | Punctilious ethyl alcohol | HMDB | | Pyro | HMDB | | Silent spirit | HMDB | | Spirit | HMDB | | Spirits OF wine | HMDB | | Spirt | HMDB | | Synasol | HMDB | | Tecsol | HMDB | | Tecsol C | HMDB | | Thanol | HMDB | | Undenatured ethanol | HMDB |
|
|---|
| Chemical Formula | C2H6O |
|---|
| Average Molecular Weight | 46.07 |
|---|
| Monoisotopic Molecular Weight | 46.0419 |
|---|
| IUPAC Name | ethanol |
|---|
| Traditional Name | ethyl alcohol |
|---|
| CAS Registry Number | 64-17-5 |
|---|
| SMILES | CCO |
|---|
| InChI Identifier | InChI=1S/C2H6O/c1-2-3/h3H,2H2,1H3 |
|---|
| InChI Key | LFQSCWFLJHTTHZ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as primary alcohols. Primary alcohols are compounds comprising the primary alcohol functional group, with the general structure RCOH (R=alkyl, aryl). |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Alcohols and polyols |
|---|
| Direct Parent | Primary alcohols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hydrocarbon derivative
- Primary alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Indirect biological role: Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | -114.1 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 1000 mg/mL | Not Available | | logP | -0.31 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-003s-9000000000-dabf5a61c5a7da9fbe13 | 2014-09-20 | View Spectrum | | GC-MS | Ethanol, non-derivatized, GC-MS Spectrum | splash10-001j-9000000000-a705823ce4aeba7f89e1 | Spectrum | | GC-MS | Ethanol, non-derivatized, GC-MS Spectrum | splash10-001j-9000000000-a705823ce4aeba7f89e1 | Spectrum | | Predicted GC-MS | Ethanol, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-002b-9000000000-7fa80a491183c1cdd23e | Spectrum | | Predicted GC-MS | Ethanol, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0fmi-9200000000-587cc3c48ab7fbf9cd3a | Spectrum | | Predicted GC-MS | Ethanol, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Ethanol, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Ethanol, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI RMU-7M) , Positive | splash10-001j-9000000000-a705823ce4aeba7f89e1 | 2012-08-31 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9000000000-d75d9996bc68c673f090 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9000000000-893c2599624722912f25 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-004i-9000000000-8af4124822065023744f | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9000000000-22231ed69c5f28bfed79 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-9000000000-d2881505c47bbef13f18 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0002-9000000000-2ed988bb761ac20ba44a | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9000000000-5c5e98353b75870c1c55 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9000000000-326f4f38d260a85ea469 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-002b-9000000000-f850ab2a079485c4a276 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9000000000-b6602db69e5662ad67a9 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-9000000000-b6602db69e5662ad67a9 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0002-9000000000-4d0d1c69f8440a7fda18 | 2021-09-24 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 90 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 25.16 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Ethanol Degradation | 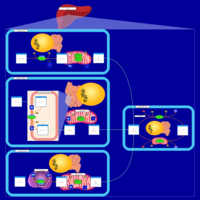 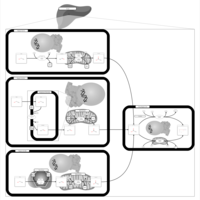  | Not Available | | Disulfiram Action Pathway | 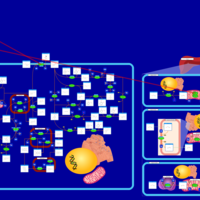   | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
| Voltage-dependent L-type calcium channel subunit alpha-1D | CACNA1D | 3p14.3 | Q01668 | details | | Voltage-dependent L-type calcium channel subunit alpha-1C | CACNA1C | 12p13.3 | Q13936 | details | | Equilibrative nucleoside transporter 1 | SLC29A1 | 6p21.1 | Q99808 | details | | Glutamate receptor 1 | GRIA1 | 5q33|5q31.1 | P42261 | details | | 5-hydroxytryptamine receptor 3A | HTR3A | 11q23.1 | P46098 | details | | Equilibrative nucleoside transporter 2 | SLC29A2 | 11q13 | Q14542 | details | | Neuronal acetylcholine receptor subunit alpha-10 | CHRNA10 | 11p15.5 | Q9GZZ6 | details | | Neuronal acetylcholine receptor subunit alpha-9 | CHRNA9 | 4p14 | Q9UGM1 | details | | Voltage-dependent L-type calcium channel subunit alpha-1S | CACNA1S | 1q32 | Q13698 | details | | Voltage-dependent L-type calcium channel subunit beta-1 | CACNB1 | 17q21-q22 | Q02641 | details | | Voltage-dependent calcium channel gamma-1 subunit | CACNG1 | 17q24 | Q06432 | details | | Voltage-dependent calcium channel gamma-2 subunit | CACNG2 | 22q13.1 | Q9Y698 | details | | Glutamate receptor 2 | GRIA2 | 4q32-q33 | P42262 | details | | Glutamate [NMDA] receptor subunit 3A | GRIN3A | 9q31.1 | Q8TCU5 | details | | Gamma-aminobutyric acid receptor subunit alpha-1 | GABRA1 | 5q34-q35 | P14867 | details | | Gamma-aminobutyric acid receptor subunit alpha-2 | GABRA2 | 4p12 | P47869 | details | | Gamma-aminobutyric acid receptor subunit alpha-3 | GABRA3 | | P34903 | details | | Gamma-aminobutyric acid receptor subunit alpha-4 | GABRA4 | 4p12 | P48169 | details | | Gamma-aminobutyric acid receptor subunit alpha-5 | GABRA5 | 15q11.2-q12 | P31644 | details | | Gamma-aminobutyric acid receptor subunit alpha-6 | GABRA6 | 5q34 | Q16445 | details | | Gamma-aminobutyric acid receptor subunit beta-1 | GABRB1 | 4p12 | P18505 | details | | Gamma-aminobutyric acid receptor subunit beta-2 | GABRB2 | 5q34 | P47870 | details | | Gamma-aminobutyric acid receptor subunit beta-3 | GABRB3 | 15q11.2-q12 | P28472 | details | | Gamma-aminobutyric acid receptor subunit delta | GABRD | 1p|1p36.3 | O14764 | details | | Gamma-aminobutyric acid receptor subunit epsilon | GABRE | | P78334 | details | | Gamma-aminobutyric acid receptor subunit gamma-1 | GABRG1 | 4p12 | Q8N1C3 | details | | Gamma-aminobutyric acid receptor subunit gamma-3 | GABRG3 | 15q12 | Q99928 | details | | Gamma-aminobutyric acid receptor subunit pi | GABRP | 5q33-q34 | O00591 | details | | Gamma-aminobutyric acid receptor subunit theta | GABRQ | | Q9UN88 | details | | Glycine receptor subunit alpha-1 | GLRA1 | 5q32 | P23415 | details | | Glycine receptor subunit alpha-2 | GLRA2 | | P23416 | details | | G protein-activated inward rectifier potassium channel 1 | KCNJ3 | 2q24.1 | P48549 | details | | G protein-activated inward rectifier potassium channel 4 | KCNJ5 | 11q24 | P48544 | details | | G protein-activated inward rectifier potassium channel 2 | KCNJ6 | 21q22.1|21q22.1 | P48051 | details | | G protein-activated inward rectifier potassium channel 3 | KCNJ9 | 1q23.2 | Q92806 | details | | Neuronal acetylcholine receptor subunit alpha-2 | CHRNA2 | 8p21 | Q15822 | details | | 5-hydroxytryptamine receptor 3B | HTR3B | 11q23.1 | O95264 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | |
| Alcohol dehydrogenase class 4 mu/sigma chain | ADH7 | 4q23-q24 | P40394 | details | | Glutamate receptor 1 | GRIA1 | 5q33|5q31.1 | P42261 | details | | Neuronal acetylcholine receptor subunit alpha-10 | CHRNA10 | 11p15.5 | Q9GZZ6 | details | | Neuronal acetylcholine receptor subunit alpha-9 | CHRNA9 | 4p14 | Q9UGM1 | details | | Glutamate receptor 2 | GRIA2 | 4q32-q33 | P42262 | details | | Glutamate [NMDA] receptor subunit 3A | GRIN3A | 9q31.1 | Q8TCU5 | details | | Gamma-aminobutyric acid receptor subunit alpha-1 | GABRA1 | 5q34-q35 | P14867 | details | | Gamma-aminobutyric acid receptor subunit alpha-2 | GABRA2 | 4p12 | P47869 | details | | Gamma-aminobutyric acid receptor subunit alpha-3 | GABRA3 | | P34903 | details | | Gamma-aminobutyric acid receptor subunit alpha-4 | GABRA4 | 4p12 | P48169 | details | | Gamma-aminobutyric acid receptor subunit alpha-5 | GABRA5 | 15q11.2-q12 | P31644 | details | | Gamma-aminobutyric acid receptor subunit alpha-6 | GABRA6 | 5q34 | Q16445 | details | | Gamma-aminobutyric acid receptor subunit beta-1 | GABRB1 | 4p12 | P18505 | details | | Gamma-aminobutyric acid receptor subunit beta-2 | GABRB2 | 5q34 | P47870 | details | | Gamma-aminobutyric acid receptor subunit beta-3 | GABRB3 | 15q11.2-q12 | P28472 | details | | Gamma-aminobutyric acid receptor subunit delta | GABRD | 1p|1p36.3 | O14764 | details | | Gamma-aminobutyric acid receptor subunit epsilon | GABRE | | P78334 | details | | Gamma-aminobutyric acid receptor subunit gamma-1 | GABRG1 | 4p12 | Q8N1C3 | details | | Gamma-aminobutyric acid receptor subunit gamma-3 | GABRG3 | 15q12 | Q99928 | details | | Gamma-aminobutyric acid receptor subunit pi | GABRP | 5q33-q34 | O00591 | details | | Gamma-aminobutyric acid receptor subunit theta | GABRQ | | Q9UN88 | details | | Glycine receptor subunit alpha-1 | GLRA1 | 5q32 | P23415 | details | | Glycine receptor subunit alpha-2 | GLRA2 | | P23416 | details | | Neuronal acetylcholine receptor subunit alpha-2 | CHRNA2 | 8p21 | Q15822 | details | | 5-hydroxytryptamine receptor 3B | HTR3B | 11q23.1 | O95264 | details | | Neuronal acetylcholine receptor subunit beta-2 | CHRNB2 | 1q21.3 | P17787 | details | | Neuronal acetylcholine receptor subunit alpha-5 | CHRNA5 | 15q24 | P30532 | details | | Neuronal acetylcholine receptor subunit beta-4 | CHRNB4 | 15q24 | P30926 | details | | Neuronal acetylcholine receptor subunit alpha-3 | CHRNA3 | 15q24 | P32297 | details | | Neuronal acetylcholine receptor subunit alpha-7 | CHRNA7 | 15q14 | P36544 | details | | Glutamate receptor 3 | GRIA3 | Xq25-q26 | P42263 | details | | Neuronal acetylcholine receptor subunit alpha-4 | CHRNA4 | 20q13.2-q13.3 | P43681 | details | | Glutamate receptor 4 | GRIA4 | 11q22 | P48058 | details | | Neuronal acetylcholine receptor subunit beta-3 | CHRNB3 | 8p11.2 | Q05901 | details | | Neuronal acetylcholine receptor subunit alpha-6 | CHRNA6 | 8p11.21 | Q15825 | details | | 5-hydroxytryptamine receptor 3D | HTR3D | | Q70Z44 | details | | 5-hydroxytryptamine receptor 3C | HTR3C | | Q8WXA8 | details | | 5-Hydroxytryptamine receptor 3E | HTR3E | | A5X5Y0 | details |
|
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000108 |
|---|
| DrugBank ID | DB00898 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB000753 |
|---|
| KNApSAcK ID | C00019560 |
|---|
| Chemspider ID | 682 |
|---|
| KEGG Compound ID | C00469 |
|---|
| BioCyc ID | ETOH |
|---|
| BiGG ID | 35062 |
|---|
| Wikipedia Link | Ethanol |
|---|
| METLIN ID | 3203 |
|---|
| PubChem Compound | 702 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16236 |
|---|
| References |
|---|
| General References | - Ryabova OB, Chmil OM, Sibirny AA: Xylose and cellobiose fermentation to ethanol by the thermotolerant methylotrophic yeast Hansenula polymorpha. FEMS Yeast Res. 2003 Nov;4(2):157-64. doi: 10.1016/S1567-1356(03)00146-6. [PubMed:14613880 ]
|
|---|