| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:25:53 UTC |
|---|
| Updated at | 2022-12-13 23:36:24 UTC |
|---|
| CannabisDB ID | CDB000130 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | L-Glutamic acid |
|---|
| Description | L-glutamic acid (abbreviated Glu or E), also referred to as glutamate (the anion), is a non-essential amino acid, one of the 20 amino acids used in the biosynthesis of proteins. L-glutamic acid exists in all living species, from bacteria to humans. In humans, dietary proteins are broken down by digestion into amino acids, which serves as metabolic fuel or other functional roles in the body. Glutamate is a component amino acid in many protein rich foods including the gluten protein in flour and is found as a free acid in cheeses and soya sauce. It is used as a flavor enhancer as a sodium salt known as monosodium glutamate. As a proteinogenic amino acid, L-glutamic acid is also found in Cannabis plants (PMID: 6991645 ). Glutamate is a key molecule in cellular metabolism. Glutamate is the most abundant fast excitatory neurotransmitter in the mammalian nervous system. At chemical synapses, glutamate is stored in vesicles. Nerve impulses trigger release of glutamate from the pre-synaptic cell. In the opposing post-synaptic cell, glutamate receptors, such as the N-methyl-d-aspartate acid (NMDA) receptor, bind glutamate and are activated. Because of its role in synaptic plasticity, it is believed that glutamic acid is involved in cognitive functions like learning and memory in the brain. Glutamate transporters are found in neuronal and glial membranes. They rapidly remove glutamate from the extracellular space. In brain injury or disease, glutamate can accumulate outside cells, causing glutamate excitotoxicity. Excitotoxicity occurs when neurons are exposed to high levels of glutamate or other neurotransmitters, causing persistent activation of the and α-amino-3-hydroxy-5-methylisoxazole propionic acid (AMPA) receptors and voltage-gated calcium channels. This results in a lethal influx of extracellular calcium leading to neuronal damage and eventual cell death. Cell death arises from damage to mitochondria by the excessively high intracellular calcium, which open mitochondrial pores, causing mitochondria to swell. Reactive oxygen species (ROS) may also be released by mitochondria into the intracellular space. Excess glutamate and calcium trigger apoptosis by further activating transcription factors for pro-apoptotic genes, or downregulating transcription factors for anti-apoptotic genes. Glutamate excitotoxicity causes other health consequences. In ischemic stroke and brain trauma, the severely reduced blood supply leads to flooding of glutamate and aspartate into the extra-neuronal space ( PMID: 16314180 ). Glutamate excitotoxicity is associated with diseases like amyotrophic lateral sclerosis, multiple sclerosis, lathyrism, and Alzheimer's disease (PMID: 20229265 ) and in epileptic seizures. Microinjection of glutamic acid into neurons produces spontaneous depolarization around one second apart which is like a paroxysmal depolarizing shift seen in epileptic attacks. This change in the resting membrane potential at seizure foci could cause spontaneous opening of voltage activated calcium channels, leading to glutamic acid release and further depolarization. Moreover, glutamic acid is associated with N-acetylglutamate synthetase deficiency, which is an inborn error of metabolism. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (S)-2-Aminopentanedioic acid | ChEBI | | (S)-Glutamic acid | ChEBI | | Acide glutamique | ChEBI | | Acido glutamico | ChEBI | | Acidum glutamicum | ChEBI | | E | ChEBI | | Glu | ChEBI | | Glutamate | ChEBI | | GLUTAMIC ACID | ChEBI | | L-Glu | ChEBI | | L-Glutaminic acid | ChEBI | | L-Glutaminsaeure | ChEBI | | (S)-2-Aminopentanedioate | Generator | | (S)-Glutamate | Generator | | L-Glutaminate | Generator | | L-Glutamate | Generator | | D-Glutamate | MeSH | | L Glutamate | MeSH | | L Glutamic acid | MeSH | | Aluminum L glutamate | MeSH | | Aluminum L-glutamate | MeSH | | Potassium glutamate | MeSH | | D Glutamate | MeSH | | Glutamate, potassium | MeSH | | Glutamic acid, (D)-isomer | MeSH | | L-Glutamate, aluminum | MeSH | | (2S)-2-Aminopentanedioate | HMDB | | (2S)-2-Aminopentanedioic acid | HMDB | | (S)-(+)-Glutamate | HMDB | | (S)-(+)-Glutamic acid | HMDB | | 1-amino-Propane-1,3-dicarboxylate | HMDB | | 1-amino-Propane-1,3-dicarboxylic acid | HMDB | | 1-Aminopropane-1,3-dicarboxylate | HMDB | | 1-Aminopropane-1,3-dicarboxylic acid | HMDB | | 2-Aminoglutarate | HMDB | | 2-Aminoglutaric acid | HMDB | | 2-Aminopentanedioate | HMDB | | 2-Aminopentanedioic acid | HMDB | | a-Aminoglutarate | HMDB | | a-Aminoglutaric acid | HMDB | | a-Glutamate | HMDB | | a-Glutamic acid | HMDB | | Aciglut | HMDB | | alpha-Aminoglutarate | HMDB | | alpha-Aminoglutaric acid | HMDB | | alpha-Glutamate | HMDB | | alpha-Glutamic acid | HMDB | | Aminoglutarate | HMDB | | Aminoglutaric acid | HMDB | | Glt | HMDB | | Glusate | HMDB | | Glut | HMDB | | Glutacid | HMDB | | Glutamicol | HMDB | | Glutamidex | HMDB | | Glutaminate | HMDB | | Glutaminic acid | HMDB | | Glutaminol | HMDB | | Glutaton | HMDB | | L-(+)-Glutamate | HMDB | | L-(+)-Glutamic acid | HMDB | | L-a-Aminoglutarate | HMDB | | L-a-Aminoglutaric acid | HMDB | | L-alpha-Aminoglutarate | HMDB | | L-alpha-Aminoglutaric acid | HMDB | | 2-Acetamido-2-deoxy-D-glucose | HMDB | | D-GlcNAc | HMDB | | N-Acetyl-D-glucosamine | HMDB | | N-Acetylchitosamine | HMDB | | N Acetyl D glucosamine | HMDB | | 2 Acetamido 2 deoxy D glucose | HMDB | | 2 Acetamido 2 deoxyglucose | HMDB | | 2-Acetamido-2-deoxyglucose | HMDB | | Acetylglucosamine | HMDB | | 3alpha,7alpha,12alpha-Trihydroxy-5beta-cholan-24-oylglycine | HMDB | | N-[(3alpha,5beta,7alpha,12alpha)-3,7,12-Trihydroxy-24-oxocholan-24-yl]glycine | HMDB | | N-Choloylglycine | HMDB | | 3a,7a,12a-Trihydroxy-5b-cholan-24-oylglycine | HMDB | | 3Α,7α,12α-trihydroxy-5β-cholan-24-oylglycine | HMDB | | N-[(3a,5b,7a,12a)-3,7,12-Trihydroxy-24-oxocholan-24-yl]glycine | HMDB | | N-[(3Α,5β,7α,12α)-3,7,12-trihydroxy-24-oxocholan-24-yl]glycine | HMDB | | Glycocholate | HMDB | | Glycine cholate | HMDB | | Glycocholic acid, sodium salt | HMDB | | Cholylglycine | HMDB | | Glycocholate sodium | HMDB | | 3alpha,7alpha,12alpha-Trihydroxy-5beta-cholanic acid-24-glycine | HMDB | | 3alpha,7alpha,12alpha-Trihydroxy-N-(carboxymethyl)-5beta-cholan-24-amide | HMDB | | 3Α,7α,12α-trihydroxy-5β-cholanic acid-24-glycine | HMDB | | 3Α,7α,12α-trihydroxy-N-(carboxymethyl)-5β-cholan-24-amide | HMDB | | Glycoreductodehydrocholic acid | HMDB | | Glycylcholate | HMDB | | Glycylcholic acid | HMDB | | N-(Carboxymethyl)-3alpha,7alpha,12alpha-trihydroxy-5beta-cholan-24-amide | HMDB | | N-(Carboxymethyl)-3α,7α,12α-trihydroxy-5β-cholan-24-amide | HMDB | | N-Choloyl-glycine | HMDB | | 3-Hydroxy-1,3,5(10)-estratrien-17-one | HMDB | | Follicular hormone | HMDB | | Folliculin | HMDB | | Oestrone | HMDB | | (+)-Estrone | HMDB | | 1,3,5(10)-Estratrien-3-ol-17-one | HMDB | | 3-Hydroxy-17-keto-estra-1,3,5-triene | HMDB | | 3-Hydroxyestra-1,3,5(10)-trien-17-one | HMDB | | 3-Hydroxyestra-1,3,5(10)-triene-17-one | HMDB | | 3-Hydroxyoestra-1,3,5(10)-trien-17-one | HMDB | | D1,3,5(10)-Estratrien-3-ol-17-one | HMDB | | Estrone, (+-)-isomer | HMDB | | Hyrex brand OF estrone | HMDB | | Estrone, (9 beta)-isomer | HMDB | | Estrovarin | HMDB | | Kestrone | HMDB | | Wehgen | HMDB | | Estrone, (8 alpha)-isomer | HMDB | | Hauck brand OF estrone | HMDB | | Unigen | HMDB | | Vortech brand OF estrone | HMDB | | alpha,beta-Hydroxypropionic acid | HMDB | | D-GroA | HMDB | | R-Glyceric acid | HMDB | | Glycerate | HMDB | | (R)-Glycerate | HMDB | | a,b-Hydroxypropionate | HMDB | | a,b-Hydroxypropionic acid | HMDB | | alpha,beta-Hydroxypropionate | HMDB | | Α,β-hydroxypropionate | HMDB | | Α,β-hydroxypropionic acid | HMDB | | R-Glycerate | HMDB | | (R)-Glyceric acid | HMDB | | D-Glycerate | HMDB | | D-Glyceric acid | HMDB | | (2R)-2,3-Dihydroxypropanoic acid | HMDB | | (R)-2,3-Dihydroxypropanoic acid | HMDB | | D-2,3-Dihydroxypropanoic acid | HMDB | | 1-Amino-2-hydroxyethane | HMDB | | 2-Amino-1-ethanol | HMDB | | 2-Amino-ethanol | HMDB | | 2-Aminoethan-1-ol | HMDB | | 2-Aminoethyl alcohol | HMDB | | 2-Hydroxyethylamine | HMDB | | Aethanolamin | HMDB | | Aminoethanol | HMDB | | beta-Aminoethanol | HMDB | | beta-Aminoethyl alcohol | HMDB | | beta-Ethanolamine | HMDB | | beta-Hydroxyethylamine | HMDB | | Colamine | HMDB | | ETA | HMDB | | Glycinol | HMDB | | Hea | HMDB | | MEA | HMDB | | MONOETHANOLAMINE | HMDB | | b-Aminoethanol | HMDB | | Β-aminoethanol | HMDB | | b-Aminoethyl alcohol | HMDB | | Β-aminoethyl alcohol | HMDB | | b-Ethanolamine | HMDB | | Β-ethanolamine | HMDB | | b-Hydroxyethylamine | HMDB | | Β-hydroxyethylamine | HMDB | | 2-Aminoethanol | HMDB | | 2-Ethanolamine | HMDB | | 2-Hydroxyethanamine | HMDB | | Envision conditioner PDD 9020 | HMDB | | Ethylolamine | HMDB | | H-Glycinol | HMDB | | Monoaethanolamin | HMDB | | Olamine | HMDB | | 2 Aminoethanol | HMDB | | (3R,4S,5R)-5-[(1R)-1-Carboxy-2,2-difluoro-1-(phosphonooxy)ethoxy]-4-hydroxy-3-(phosphonooxy)cyclohex-1-ene-1-carboxylate | HMDB | | (1S)-2-[(3-O-b-D-Glucopyranosyl-b-D-galactopyranosyl)oxy]-1-{[(9E)-octadec-9-enoyloxy]methyl}ethyl (10E)-nonadec-10-enoate | HMDB | | (1S)-2-[(3-O-b-D-Glucopyranosyl-b-D-galactopyranosyl)oxy]-1-{[(9E)-octadec-9-enoyloxy]methyl}ethyl (10E)-nonadec-10-enoic acid | HMDB | | (1S)-2-[(3-O-beta-D-Glucopyranosyl-beta-D-galactopyranosyl)oxy]-1-{[(9E)-octadec-9-enoyloxy]methyl}ethyl (10E)-nonadec-10-enoic acid | HMDB | | (1S)-2-[(3-O-Β-D-glucopyranosyl-β-D-galactopyranosyl)oxy]-1-{[(9E)-octadec-9-enoyloxy]methyl}ethyl (10E)-nonadec-10-enoate | HMDB | | (1S)-2-[(3-O-Β-D-glucopyranosyl-β-D-galactopyranosyl)oxy]-1-{[(9E)-octadec-9-enoyloxy]methyl}ethyl (10E)-nonadec-10-enoic acid | HMDB |
|
|---|
| Chemical Formula | C5H9NO4 |
|---|
| Average Molecular Weight | 147.13 |
|---|
| Monoisotopic Molecular Weight | 147.0532 |
|---|
| IUPAC Name | (2S)-2-aminopentanedioic acid |
|---|
| Traditional Name | L-glutamic acid |
|---|
| CAS Registry Number | 56-86-0 |
|---|
| SMILES | N[C@@H](CCC(O)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C5H9NO4/c6-3(5(9)10)1-2-4(7)8/h3H,1-2,6H2,(H,7,8)(H,9,10)/t3-/m0/s1 |
|---|
| InChI Key | WHUUTDBJXJRKMK-VKHMYHEASA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as glutamic acid and derivatives. Glutamic acid and derivatives are compounds containing glutamic acid or a derivative thereof resulting from reaction of glutamic acid at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Glutamic acid and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glutamic acid or derivatives
- Alpha-amino acid
- L-alpha-amino acid
- Amino fatty acid
- Dicarboxylic acid or derivatives
- Fatty acid
- Fatty acyl
- Amino acid
- Carboxylic acid
- Organic oxide
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Organopnictogen compound
- Carbonyl group
- Organic oxygen compound
- Amine
- Organic nitrogen compound
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Biological role: Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 8.57 mg/mL | Not Available | | logP | -3.69 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | L-Glutamic acid, 3 TMS, GC-MS Spectrum | splash10-0002-2950000000-2d6edc93ec5f8dee2223 | Spectrum | | GC-MS | L-Glutamic acid, non-derivatized, GC-MS Spectrum | splash10-002b-0940000000-4e285988bc537825d94d | Spectrum | | GC-MS | L-Glutamic acid, 3 TMS, GC-MS Spectrum | splash10-00dj-9630000000-1ecc76aab86283892139 | Spectrum | | GC-MS | L-Glutamic acid, 2 TMS, GC-MS Spectrum | splash10-001i-8910000000-00f65ced0c55aa4ad169 | Spectrum | | GC-MS | L-Glutamic acid, 3 TMS, GC-MS Spectrum | splash10-0032-3980000000-3069de5b6c49e4176968 | Spectrum | | GC-MS | L-Glutamic acid, non-derivatized, GC-MS Spectrum | splash10-0002-0790000000-79d3e289c22cb183faa1 | Spectrum | | GC-MS | L-Glutamic acid, non-derivatized, GC-MS Spectrum | splash10-0002-2950000000-2d6edc93ec5f8dee2223 | Spectrum | | GC-MS | L-Glutamic acid, non-derivatized, GC-MS Spectrum | splash10-002b-0940000000-4e285988bc537825d94d | Spectrum | | GC-MS | L-Glutamic acid, non-derivatized, GC-MS Spectrum | splash10-00ea-6932100000-30d3f5dcc198a5971e96 | Spectrum | | GC-MS | L-Glutamic acid, non-derivatized, GC-MS Spectrum | splash10-00dj-9630000000-1ecc76aab86283892139 | Spectrum | | GC-MS | L-Glutamic acid, non-derivatized, GC-MS Spectrum | splash10-001i-8910000000-00f65ced0c55aa4ad169 | Spectrum | | GC-MS | L-Glutamic acid, non-derivatized, GC-MS Spectrum | splash10-0032-3980000000-3069de5b6c49e4176968 | Spectrum | | Predicted GC-MS | L-Glutamic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0znc-9300000000-f88e86b78a4cee99a2d4 | Spectrum | | Predicted GC-MS | L-Glutamic acid, 2 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00di-8290000000-f99e03763f636e557887 | Spectrum | | Predicted GC-MS | L-Glutamic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Glutamic acid, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Glutamic acid, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Glutamic acid, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Glutamic acid, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Glutamic acid, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Glutamic acid, TMS_2_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Glutamic acid, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Glutamic acid, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Glutamic acid, TBDMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Glutamic acid, TBDMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-003r-6900000000-95b0a084dc076f9c7b91 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001i-9000000000-c37d4c80a14ec029ef63 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0a59-9000000000-6f1888aa71bcb0adf76c | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-004j-0900000000-5fa8a338dcd2f2a6bdd2 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-004i-0900000000-16763200aa07f7629ad4 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-03di-3900000000-d9cfc5187aa799f6f978 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0a4i-0900000000-10ee9a593e13550bec1c | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0002-0900000000-4d045a3c1fc6e56f801b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-9000000000-c3c7f8f3754109a0c25b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-004i-0900000000-48bfae26c69408b7f0ae | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0002-0900000000-82c2a681e7522a7bb1d1 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0002-0905010000-af1c9ec4d0062fa6960e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-01q9-9700000000-2b967b896a6e48914512 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-0900000000-1434321646181ea894a0 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-0900000000-11fadb2530828eedad8a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-006t-0941100000-07d051890cb1d9e5c856 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-9200000000-f8619d11f1f54d836bb1 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-0900000000-ea2dd00e79ef06e8dc04 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-0900000000-3e239d4014c95a2ef873 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0002-0900000000-b548959edce39d319cf4 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0udi-0900000000-40d901f655797db2cd0f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0udi-1900000000-f997527a39900ac431c7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0udi-7900000000-60816f0601a4e6d25ffb | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0007-9000000000-6ee821f657c604d1afca | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-000t-0900000000-7c02624abe56da9247a2 | 2012-08-31 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Alanine Metabolism | 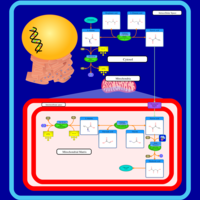   |  | | Histidine Metabolism | 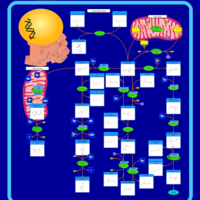  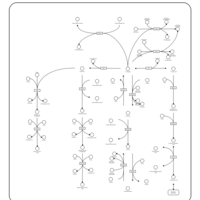 |  | | Malate-Aspartate Shuttle |  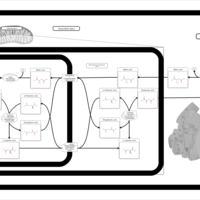 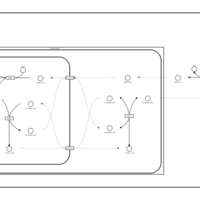 | Not Available | | Ammonia Recycling |  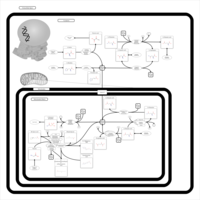 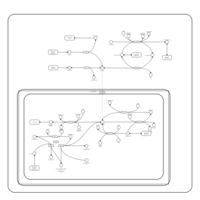 |  | | Cysteine Metabolism | 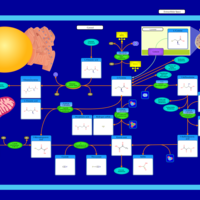 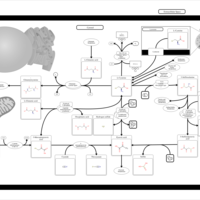 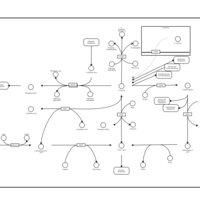 |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial | ALDH4A1 | 1p36 | P30038 | details | | Glutathione reductase, mitochondrial | GSR | 8p21.1 | P00390 | details | | Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 2 | GFPT2 | 5q34-q35 | O94808 | details | | Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | GFPT1 | 2p13 | Q06210 | details | | 4-aminobutyrate aminotransferase, mitochondrial | ABAT | 16p13.2 | P80404 | details | | Tyrosine aminotransferase | TAT | 16q22.1 | P17735 | details | | Aspartate aminotransferase, cytoplasmic | GOT1 | 10q24.1-q25.1 | P17174 | details | | Aspartate aminotransferase, mitochondrial | GOT2 | 16q21 | P00505 | details | | Branched-chain-amino-acid aminotransferase, cytosolic | BCAT1 | 12p12.1 | P54687 | details | | Amidophosphoribosyltransferase | PPAT | 4q12 | Q06203 | details | | Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | AADAT | 4q33 | Q8N5Z0 | details | | Alanine--glyoxylate aminotransferase 2, mitochondrial | AGXT2 | 5p13 | Q9BYV1 | details | | Ornithine aminotransferase, mitochondrial | OAT | 10q26 | P04181 | details | | Kynurenine--oxoglutarate transaminase 1 | CCBL1 | 9q34.11 | Q16773 | details | | Glutamate--cysteine ligase catalytic subunit | GCLC | 6p12 | P48506 | details | | Glutamate--cysteine ligase regulatory subunit | GCLM | 1p22.1 | P48507 | details | | N-acetylglutamate synthase, mitochondrial | NAGS | 17q21.31 | Q8N159 | details | | Branched-chain-amino-acid aminotransferase, mitochondrial | BCAT2 | 19q13 | O15382 | details | | Bifunctional glutamate/proline--tRNA ligase | EPRS | 1q41 | P07814 | details | | Asparagine synthetase [glutamine-hydrolyzing] | ASNS | 7q21.3 | P08243 | details | | GMP synthase [glutamine-hydrolyzing] | GMPS | 3q24 | P49915 | details | | Glutamate dehydrogenase 2, mitochondrial | GLUD2 | Xq24-q25 | P49448 | details | | Glutamine synthetase | GLUL | 1q31 | P15104 | details | | Glutaminase liver isoform, mitochondrial | GLS2 | 12q13 | Q9UI32 | details | | CTP synthase 1 | CTPS1 | 1p34.1 | P17812 | details | | Glutaminase kidney isoform, mitochondrial | GLS | 2q32-q34 | O94925 | details | | Formimidoyltransferase-cyclodeaminase | FTCD | 21q22.3 | O95954 | details | | Glutamate dehydrogenase 1, mitochondrial | GLUD1 | 10q23.3 | P00367 | details | | Glutamate decarboxylase 2 | GAD2 | 10p11.23 | Q05329 | details | | Glutamate decarboxylase 1 | GAD1 | 2q31 | Q99259 | details | | Gamma-glutamyltranspeptidase 1 | GGT1 | 22q11.23 | P19440 | details | | Gamma-glutamyltransferase 7 | GGT7 | 20q11.22 | Q9UJ14 | details | | Serine--pyruvate aminotransferase | AGXT | 2q37.3 | P21549 | details | | Glutathione synthetase | GSS | 20q11.2 | P48637 | details | | Folylpolyglutamate synthase, mitochondrial | FPGS | 9q34.1 | Q05932 | details | | Alanine aminotransferase 1 | GPT | 8q24.3 | P24298 | details | | Phosphoserine aminotransferase | PSAT1 | 9q21.2 | Q9Y617 | details | | Alpha-aminoadipic semialdehyde synthase, mitochondrial | AASS | 7q31.3 | Q9UDR5 | details | | Phosphoribosylformylglycinamidine synthase | PFAS | 17p13.1 | O15067 | details | | Delta-1-pyrroline-5-carboxylate synthase | ALDH18A1 | 10q24.3 | P54886 | details | | 5-oxoprolinase | OPLAH | 8q24.3 | O14841 | details | | Vitamin K-dependent gamma-carboxylase | GGCX | 2p12 | P38435 | details | | Glutamate receptor, ionotropic kainate 1 | GRIK1 | 21q22.11 | P39086 | details | | Glutamate receptor, ionotropic kainate 3 | GRIK3 | 1p34-p33 | Q13003 | details | | Glutamyl-tRNA(Gln) amidotransferase subunit B, mitochondrial | PET112 | 4q31.3 | O75879 | details | | Glutamate carboxypeptidase 2 | FOLH1 | 11p11.2 | Q04609 | details | | Arginine decarboxylase | ADC | 1p35.1 | Q96A70 | details | | Glutamate receptor 1 | GRIA1 | 5q33|5q31.1 | P42261 | details | | Aminoadipate aminotransferase, isoform CRA_b | AADAT | 4q33 | Q4W5N8 | details | | Mitochondrial glutamate carrier 2 | SLC25A18 | 22q11.2 | Q9H1K4 | details | | Glutamine-dependent NAD(+) synthetase | NADSYN1 | 11q13.4 | Q6IA69 | details | | Alanine aminotransferase 2 | GPT2 | 16q12.1 | Q8TD30 | details | | Lengsin | LGSN | 6pter-q22.33 | Q5TDP6 | details | | CTP synthase 2 | CTPS2 | Xp22 | Q9NRF8 | details | | Mitochondrial glutamate carrier 1 | SLC25A22 | 11p15.5 | Q9H936 | details | | Gamma-glutamyltransferase 6 | GGT6 | 17p13.2 | Q6P531 | details | | Probable glutamate--tRNA ligase, mitochondrial | EARS2 | 16p12.2 | Q5JPH6 | details | | Aminoadipate-semialdehyde synthase | AASS | 7q31.3 | A4D0W4 | details | | N-acetylaspartyl-glutamate synthetase A | RIMKLA | 1p34.2 | Q8IXN7 | details | | Beta-citryl-glutamate synthase B | RIMKLB | 12p13.31 | Q9ULI2 | details | | Putative aspartate aminotransferase, cytoplasmic 2 | GOT1L1 | 8p11.23 | Q8NHS2 | details | | Kynurenine--oxoglutarate transaminase 3 | CCBL2 | 1p22.2 | Q6YP21 | details | | Aspartate aminotransferase | GIG18 | | Q2TU84 | details | | GGT7 protein | GGT7 | 20q11.22 | A0PJJ9 | details | | Gamma-glutamyltransferase 5 | GGT5 | 22q11.23 | P36269 | details | | Glutamyl aminopeptidase | ENPEP | 4q25 | Q07075 | details | | Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial | QRSL1 | 6q21 | Q9H0R6 | details | | Aspartyl aminopeptidase | DNPEP | 2q35 | Q9ULA0 | details | | Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial | GATC | 12q24.31 | O43716 | details | | Gamma-glutamyltranspeptidase 2 | GGT2 | | P36268 | details | | Putative gamma-glutamyltranspeptidase 3 | GGT3P | | A6NGU5 | details |
|
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | |
| Glutamate--cysteine ligase catalytic subunit | GCLC | 6p12 | P48506 | details |
|
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 0.516 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 0.521 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 0.225 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.552 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 0.222 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 0.259 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000148 |
|---|
| DrugBank ID | DB00142 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB012535 |
|---|
| KNApSAcK ID | C00001358 |
|---|
| Chemspider ID | Not Available |
|---|
| KEGG Compound ID | C00025 |
|---|
| BioCyc ID | GLT |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | L-Glutamic_Acid |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 33032 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16015 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
- Camacho A, Massieu L: Role of glutamate transporters in the clearance and release of glutamate during ischemia and its relation to neuronal death. Arch Med Res. 2006 Jan;37(1):11-8. doi: 10.1016/j.arcmed.2005.05.014. [PubMed:16314180 ]
- Lau A, Tymianski M: Glutamate receptors, neurotoxicity and neurodegeneration. Pflugers Arch. 2010 Jul;460(2):525-42. doi: 10.1007/s00424-010-0809-1. Epub 2010 Mar 14. [PubMed:20229265 ]
|
|---|



