| Identification |
|---|
| HMDB Protein ID
| CDBP02311 |
| Secondary Accession Numbers
| Not Available |
| Name
| Glutamine-dependent NAD(+) synthetase |
| Description
| Not Available |
| Synonyms
|
- NAD(+) synthase [glutamine-hydrolyzing]
- NAD(+) synthetase
|
| Gene Name
| NADSYN1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in NAD+ synthase (glutamine-hydrolyzing) activity |
| Specific Function
| Not Available |
| GO Classification
|
| Biological Process |
| NAD biosynthetic process |
| water-soluble vitamin metabolic process |
| NAD metabolic process |
| Cellular Component |
| cytosol |
| Function |
| nad+ synthase (glutamine-hydrolyzing) activity |
| binding |
| catalytic activity |
| hydrolase activity |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds |
| nucleoside binding |
| purine nucleoside binding |
| adenyl nucleotide binding |
| adenyl ribonucleotide binding |
| atp binding |
| ligase activity |
| carbon-nitrogen ligase activity, with glutamine as amido-n-donor |
| ligase activity, forming carbon-nitrogen bonds |
| Molecular Function |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds |
| NAD+ synthase (glutamine-hydrolyzing) activity |
| ATP binding |
| Process |
| metabolic process |
| nitrogen compound metabolic process |
| coenzyme biosynthetic process |
| pyridine nucleotide biosynthetic process |
| nicotinamide nucleotide biosynthetic process |
| nad biosynthetic process |
| cellular metabolic process |
| cofactor metabolic process |
| coenzyme metabolic process |
|
| Cellular Location
|
- Cytoplasmic
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Glutamate Metabolism | 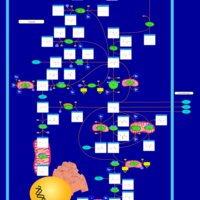 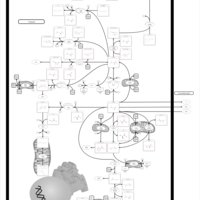 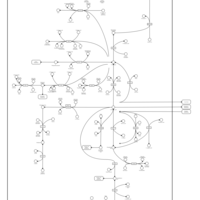 |  | | Nicotinate and Nicotinamide Metabolism | 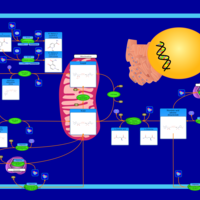 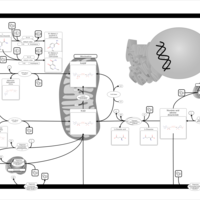 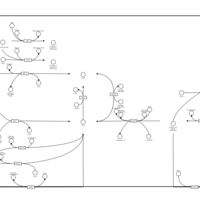 |  | | NAD(+) biosynthesis | Not Available | Not Available | | 4-Hydroxybutyric Aciduria/Succinic Semialdehyde Dehydrogenase Deficiency | 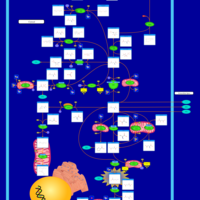 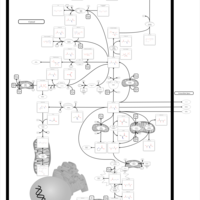 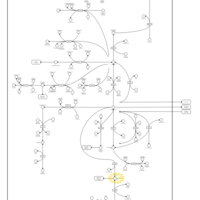 | Not Available | | Homocarnosinosis | 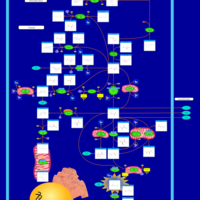 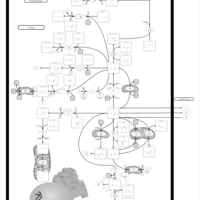 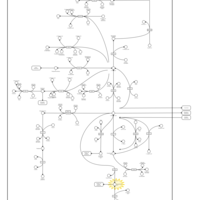 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 11 |
| Locus
| 11q13.4 |
| SNPs
| NADSYN1 |
| Gene Sequence
|
>2121 bp
ATGGGCCGGAAGGTGACCGTGGCCACCTGCGCACTCAACCAGTGGGCCCTGGACTTCGAG
GGCAATTTGCAAAGAATTTTAAAGAGTATTGAAATTGCCAAAAACAGAGGAGCAAGATAC
AGGCTTGGACCAGAGCTGGAAATATGCGGCTACGGATGTTGGGATCATTATTACGAGTCG
GACACCCTCTTGCACTCGTTTCAAGTCCTAGCGGCCCTTGTGGAGTCTCCCGTCACTCAG
GACATCATCTGCGACGTGGGGATGCCTGTAATGCACCGAAACGTCCGCTACAACTGCAGA
GTGGTATTCCTCAACAGGAAGATCCTGCTCATCAGACCCAAGATGGCCTTGGCCAATGAA
GGCAACTACCGCGAGCTGCGCTGGTTCACCCCGTGGTCGAGGAGTCGGCACACAGAGGAG
TACTTTCTGCCTCGGATGATACAGGACCTGACAAAGCAGGAAACCGTACCCTTCGGAGAT
GCGGTGCTGGTGACATGGGACACCTGCATTGGAAGTGAGATCTGTGAGGAGCTCTGGACA
CCCCACAGCCCGCACATCGACATGGGCCTGGATGGCGTGGAGATCATCACCAACGCCTCG
GGCAGCCACCACGTGCTGCGCAAAGCCAACACCAGGGTGGATCTCGTGACTATGGTCACC
AGCAAGAACGGTGGGATTTACTTGCTGGCCAACCAGAAGGGTTGCGACGGGGACCGCCTG
TACTACGACGGCTGTGCCATGATCGCCATGAACGGAAGCGTCTTTGCTCAAGGATCCCAG
TTTTCTCTGGATGACGTGGAAGTCCTGACGGCCACGCTGGATCTGGAGGACGTCCGGAGC
TACAGGGCGGAGATTTCATCTCGAAACCTGGCGGCCAGCAGGGCGAGCCCCTACCCCAGA
GTGAAGGTGGACTTTGCCCTCTCGTGCCACGAGGACTTGCTGGCACCCATCTCTGAGCCC
ATCGAGTGGAAATACCACAGCCCTGAGGAGGAGATAAGCCTTGGACCTGCCTGCTGGCTC
TGGGATTTTTTAAGACGAAGTCAACAGGCAGGGTTTTTGCTGCCCTTGAGTGGCGGGGTG
GACAGCGCAGCCACCGCCTGCCTCATCTACTCCATGTGCTGCCAGGTCTGCGAGGCCGTG
AGGAGTGGAAATGAGGAAGTGCTGGCTGATGTCCGCACCATCGTGAACCAGATCAGCTAC
ACCCCCCAGGATCCCCGAGACCTCTGTGGACGCATACTGACCACCTGCTACATGGCCAGC
AAGAACTCCTCCCAGGAGACGTGCACCCGGGCCAGAGAGTTGGCCCAGCAGATTGGAAGC
CACCACATCAGTCTCAACATCGATCCAGCCGTGAAGGCCGTCATGGGCATCTTCAGCCTG
GTGACGGGGAAGAGCCCTCTGTTTGCAGCTCATGGAGGAAGCAGCAGGGAAAACCTGGCG
CTGCAAAATGTGCAGGCTCGAATACGGATGGTCCTCGCCTATCTGTTTGCTCAGTTGAGC
CTCTGGTCTCGGGGTGTCCACGGTGGGCTCCTCGTGCTGGGATCCGCCAACGTGGATGAG
AGTCTCCTGGGCTACCTGACCAAGTACGACTGCTCCAGTGCGGACATCAACCCCATAGGC
GGGATCAGCAAGACGGACCTCAGGGCCTTCGTCCAGTTCTGCATCCAGCGCTTCCAGCTT
CCTGCCCTGCAGAGCATCCTGTTGGCGCCGGCCACCGCAGAGCTGGAGCCCTTGGCTGAT
GGACAGGTGTCCCAGACCGACGAGGAAGATATGGGGATGACATATGCGGAGCTCTCGGTC
TATGGGAAACTCAGGAAGGTGGCCAAGATGGGGCCCTACAGCATGTTCTGCAAACTCCTC
GGCATGCGGAGACACATCTGCACCCCGAGACAGGTCGCTGACAAAGTGAAGCGGTTTTTC
TCCAAGTACTCCATGAACAGACACAAGATGACCACGCTCACACCCGCGTACCACGCCGAG
AACTACAGCCCTGAGGACAACAGGTTTGATCTGCGACCATTTCTGTACAACACAAGCTGG
CCTTGGCAGTTTCGGTGCATAGAAAATCAGGTGCTACAGCTCGAGAGGGCAGAGCCACAG
TCCCTGGACGGCGTGGACTGA
|
| Protein Properties |
|---|
| Number of Residues
| 706 |
| Molecular Weight
| 79283.945 |
| Theoretical pI
| 6.442 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Glutamine-dependent NAD(+) synthetase
MGRKVTVATCALNQWALDFEGNLQRILKSIEIAKNRGARYRLGPELEICGYGCWDHYYES
DTLLHSFQVLAALVESPVTQDIICDVGMPVMHRNVRYNCRVIFLNRKILLIRPKMALANE
GNYRELRWFTPWSRSRHTEEYFLPRMIQDLTKQETVPFGDAVLVTWDTCIGSEICEELWT
PHSPHIDMGLDGVEIITNASGSHQVLRKANTRVDLVTMVTSKNGGIYLLANQKGCDGDRL
YYDGCAMIAMNGSVFAQGSQFSLDDVEVLTATLDLEDVRSYRAEISSRNLAASRASPYPR
VKVDFALSCHEDLLAPISEPIEWKYHSPEEEISLGPACWLWDFLRRSQQAGFLLPLSGGV
DSAATACLIYSMCCQVCEAVRSGNEEVLADVRTIVNQISYTPQDPRDLCGRILTTCYMAS
KNSSQETCTRARELAQQIGSHHISLNIDPAVKAVMGIFSLVTGKSPLFAAHGGSSRENLA
LQNVQARIRMVLAYLFAQLSLWSRGVHGGLLVLGSANVDESLLGYLTKYDCSSADINPIG
GISKTDLRAFVQFCIQRFQLPALQSILLAPATAELEPLADGQVSQTDEEDMGMTYAELSV
YGKLRKVAKMGPYSMFCKLLGMWRHICTPRQVADKVKRFFSKYSMNRHKMTTLTPAYHAE
NYSPEDNRFDLRPFLYNTSWPWQFRCIENQVLQLERAEPQSLDGVD
|
| External Links |
|---|
| GenBank ID Protein
| 28849201 |
| UniProtKB/Swiss-Prot ID
| Q6IA69 |
| UniProtKB/Swiss-Prot Entry Name
| NADE_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| AB091316 |
| GeneCard ID
| NADSYN1 |
| GenAtlas ID
| NADSYN1 |
| HGNC ID
| HGNC:29832 |
| References |
|---|
| General References
| Not Available |

