| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:21:24 UTC |
|---|
| Updated at | 2022-12-13 23:36:27 UTC |
|---|
| CannabisDB ID | CDB006138 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Succinic acid |
|---|
| Description | Succinic acid is a four-carbon acyclic dicarboxylic acid. It is a white, odorless solid with a highly acidic taste. It is used as a flavoring agent, contributing a sour and astringent component characteristic of the umami taste (PMID:21932253 ). The anion, succinate, is a key component of the citric acid or TCA cycle and is capable of donating electrons to the electron transfer chain. Succinate dehydrogenase (SDH) plays an important role in mitochondrial function, being both part of the respiratory chain and the Krebs cycle. SDH, with a covalently attached FAD prosthetic group, is able to bind several different enzyme substrates (succinate and fumarate) and physiological regulators (oxaloacetate and ATP). Oxidizing succinate links SDH to the fast-cycling Krebs cycle portion where it participates in the breakdown of acetyl-CoA throughout the entire Krebs cycle. Succinate can readily be imported into the mitochondrial matrix by the n-butyl malonate- (or phenyl succinate-) sensitive dicarboxylate carrier in exchange with inorganic phosphate or another organic acid, e. g. malate (PMID:16143825 ). Human mutations in the four genes encoding the subunits of the mitochondrial succinate dehydrogenase are associated with a wide spectrum of clinical presentations, i.e.: Huntington’s disease (PMID:11803021 ). Moreover, succinic acid is found to be associated with D-2-hydroxyglutaric aciduria, which is an inborn error of metabolism. Additionally, succinic acid has been identified as an oncometabolite or an endogenous cancer-causing metabolite. Oncoetabolites are metabolic intermediates whose accumulation causes a metabolic and non-metabolic dysregulation leading to tumorigenesis. (PMID:23999438 , PMID:27117029 ). High levels of succinic acid can be found in tumors or biofluids surrounding tumors. Its oncogenic action appears due to its ability to inhibit prolyl hydroxylase domain-containing enzymes. In many tumours, oxygen availability becomes limited (hypoxia) very quickly due to rapid cell proliferation and limited blood vessel growth. The major regulator of the response to hypoxia is the HIF transcription factor (HIF-alpha). Under normal oxygen levels, protein levels of HIF-alpha are very low due to constant degradation, mediated by a series of post-translational modification events catalyzed by the prolyl hydroxylase domain-containing enzymes PHD1, 2 and 3, (also known as EglN2, 1 and 3) that hydroxylate HIF-alpha and lead to its degradation. All three of the PHD enzymes are inhibited by succinate. Succinic acid has been found to be associated with D-2-hydroxyglutaric aciduria, which is an inborn error of metabolism. Succinic acid is also a microbial metabolite. Indeed, urinary succinic acid is produced by Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumonia, Enterobacter sp., Acinetobacter sp., Proteus mirabilis, Citrobactes frundii, Enterococcus faecalis (PMID: 22292465 ). Succinic acid is also found in Actinobacillus, Anaerobiospirillum, Mannheimia, Corynebacterium and Basfia (PMID: 22292465 ; PMID: 18191255 ; PMID: 26360870 ). Succinic acid, or its anion succinate, is used as an excipient in pharmaceutical products to control acidity or as a counter ion. Drugs involving succinate include metoprolol succinate, sumatriptan succinate, Doxylamine succinate or solifenacin succinate. In 2004, succinic acid was identified by the Department of Energy of the United States of America as one of twelve molecules that can be produced from plant sugars through biological or chemical processes and that have a potential to subsequently be converted to a number of high-value bio-based chemicals or materials. In this regard, industrial hemp (Cannabis sativa L.) has been identified as a source of succinic acid after several chemical and biological treatments (PMID:25682224 ), suggessting it could serve as a promising source of building blocks for green chemistry products. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,2-Ethanedicarboxylic acid | ChEBI | | Acide butanedioique | ChEBI | | Acide succinique | ChEBI | | Acidum succinicum | ChEBI | | Amber acid | ChEBI | | Asuccin | ChEBI | | Bernsteinsaeure | ChEBI | | Butandisaeure | ChEBI | | Butanedionic acid | ChEBI | | Dihydrofumaric acid | ChEBI | | e363 | ChEBI | | Ethylenesuccinic acid | ChEBI | | HOOC-CH2-CH2-COOH | ChEBI | | Spirit OF amber | ChEBI | | 1,2-Ethanedicarboxylate | Generator | | Butanedionate | Generator | | Dihydrofumarate | Generator | | Ethylenesuccinate | Generator | | Succinate | Generator | | 1,2 Ethanedicarboxylic acid | MeSH | | 1,4 Butanedioic acid | MeSH | | 1,4-Butanedioic acid | MeSH | | Ammonium succinate | MeSH | | Butanedioic acid | MeSH | | Potassium succinate | MeSH | | Succinate, ammonium | MeSH | | Succinate, potassium | MeSH | | 1,4-Butanedioate | HMDB | | Katasuccin | HMDB | | Wormwood acid | HMDB | | 2-Acetamido-2-deoxy-D-glucose | ChEBI, HMDB | | D-GlcNAc | ChEBI, HMDB | | N-Acetyl-D-glucosamine | ChEBI, HMDB | | N-Acetylchitosamine | ChEBI, HMDB | | N Acetyl D glucosamine | HMDB | | 2 Acetamido 2 deoxy D glucose | HMDB | | 2 Acetamido 2 deoxyglucose | HMDB | | 2-Acetamido-2-deoxyglucose | HMDB | | Acetylglucosamine | HMDB |
|
|---|
| Chemical Formula | C4H6O4 |
|---|
| Average Molecular Weight | 118.09 |
|---|
| Monoisotopic Molecular Weight | 118.0266 |
|---|
| IUPAC Name | butanedioic acid |
|---|
| Traditional Name | succinic acid |
|---|
| CAS Registry Number | 110-15-6 |
|---|
| SMILES | OC(=O)CCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C4H6O4/c5-3(6)1-2-4(7)8/h1-2H2,(H,5,6)(H,7,8) |
|---|
| InChI Key | KDYFGRWQOYBRFD-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as dicarboxylic acids and derivatives. These are organic compounds containing exactly two carboxylic acid groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Dicarboxylic acids and derivatives |
|---|
| Direct Parent | Dicarboxylic acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Fatty acid
- Dicarboxylic acid or derivatives
- Carboxylic acid
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Indirect biological role: Biological role: Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 185 - 188 °C | Not Available | | Boiling Point | 235 °C | Wikipedia | | Water Solubility | 83.2 mg/mL | Not Available | | logP | -0.59 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-05di-9100000000-c629bea41d0d3d896425 | 2014-09-20 | View Spectrum | | GC-MS | Succinic acid, 2 TMS, GC-MS Spectrum | splash10-0002-0920000000-f286e6204a4163b823ba | Spectrum | | GC-MS | Succinic acid, non-derivatized, GC-MS Spectrum | splash10-0002-0900000000-bf336910bb37d7f78140 | Spectrum | | GC-MS | Succinic acid, 2 TMS, GC-MS Spectrum | splash10-006t-9800000000-df5ff4e8457d2d4ef919 | Spectrum | | GC-MS | Succinic acid, 2 TMS, GC-MS Spectrum | splash10-00c1-3930000000-3cc18e719822b5af661a | Spectrum | | GC-MS | Succinic acid, non-derivatized, GC-MS Spectrum | splash10-004i-9000000000-93b4807ae6275a3e59d7 | Spectrum | | GC-MS | Succinic acid, non-derivatized, GC-MS Spectrum | splash10-0kos-9100000000-f1df0903a24c305e68ec | Spectrum | | GC-MS | Succinic acid, non-derivatized, GC-MS Spectrum | splash10-0002-0910000000-300c33b39fb991b5a73e | Spectrum | | GC-MS | Succinic acid, non-derivatized, GC-MS Spectrum | splash10-0002-0920000000-f286e6204a4163b823ba | Spectrum | | GC-MS | Succinic acid, non-derivatized, GC-MS Spectrum | splash10-0002-0900000000-bf336910bb37d7f78140 | Spectrum | | GC-MS | Succinic acid, non-derivatized, GC-MS Spectrum | splash10-006t-9800000000-df5ff4e8457d2d4ef919 | Spectrum | | GC-MS | Succinic acid, non-derivatized, GC-MS Spectrum | splash10-00c1-3930000000-3cc18e719822b5af661a | Spectrum | | GC-MS | Succinic acid, non-derivatized, GC-MS Spectrum | splash10-0002-0900000000-f838d863ee7c2b111f02 | Spectrum | | Predicted GC-MS | Succinic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00xr-9300000000-f0644daf4fbb11fcc2dc | Spectrum | | Predicted GC-MS | Succinic acid, 2 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00dj-9710000000-ff8325384b9eefd19106 | Spectrum | | Predicted GC-MS | Succinic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Succinic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Succinic acid, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Succinic acid, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Succinic acid, TBDMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-00di-9300000000-f9dc864d93a09d3074f9 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-00di-9300000000-76c151de384928b2256f | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-01b9-7900000000-51d2341c097f04827944 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (Unknown) , Positive | splash10-004i-9000000000-93b4807ae6275a3e59d7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-03dj-0971010000-37d214dc7a8fdc26116b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-014i-9000000000-249222ac742c1634cec9 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-9000000000-6897d49472dba6a34a27 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0udi-0490000000-d138f8023125921b4b82 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-014i-1900000000-4ffdabe5bde527b66982 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-00di-9100000000-c20baa818f5ff5f678c1 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-00di-9000000000-7a49a18aa6fcb2540a12 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-00di-9000000000-9955aeb0e5a9f88ae70e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-00di-9000000000-7e1f195f111b4eafb4fa | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-00xr-9400000000-e50afc90e20cd420ba9b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-00xr-9600000000-43167f2549cbb5d5f7e8 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-014i-1900000000-4ffdabe5bde527b66982 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-00di-9100000000-7a8bfa543dc087bea06d | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-00di-9000000000-fdec6c7458176f3cbeb8 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-00di-9000000000-9955aeb0e5a9f88ae70e | 2017-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0gb9-2900000000-9d959a53833b07094158 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0l6r-9600000000-c367e11e737714d41418 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-056r-9000000000-e65aa602a8293debec36 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-2900000000-e1f840494c9003279869 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-7900000000-1089efd4a3469bcf14f1 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0604-9000000000-0e0e60bb202ffb004894 | 2016-09-12 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 22.53 MHz, DMSO-d6, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Citric Acid Cycle | 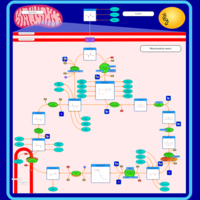 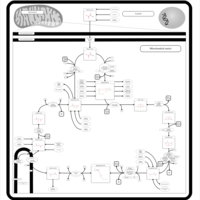 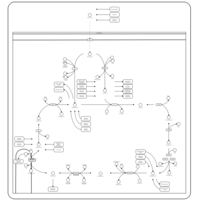 |  | | Glutamate Metabolism | 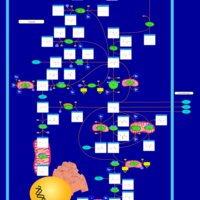 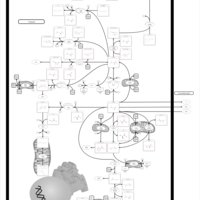 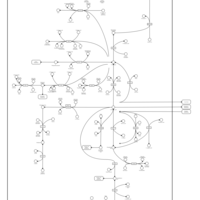 |  | | Mitochondrial Electron Transport Chain | 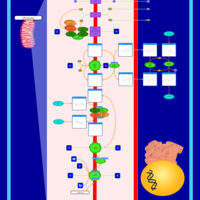 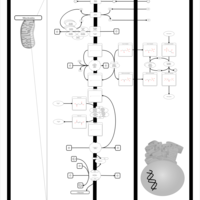 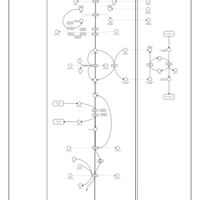 |  | | Phytanic Acid Peroxisomal Oxidation |  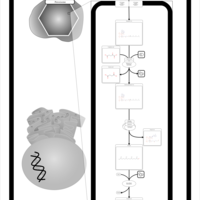 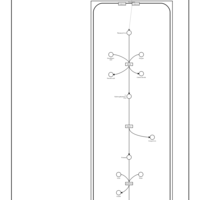 | Not Available | | Carnitine Synthesis |   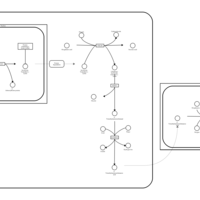 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Succinate dehydrogenase cytochrome b560 subunit, mitochondrial | SDHC | 1q23.3 | Q99643 | details | | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | SDHA | 5p15 | P31040 | details | | Succinate dehydrogenase [ubiquinone] cytochrome b small subunit, mitochondrial | SDHD | 11q23 | O14521 | details | | Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial | SDHB | 1p36.1-p35 | P21912 | details | | Succinate-semialdehyde dehydrogenase, mitochondrial | ALDH5A1 | 6p22 | P51649 | details | | Phytanoyl-CoA dioxygenase, peroxisomal | PHYH | 10p13 | O14832 | details | | Prolyl 4-hydroxylase subunit alpha-2 | P4HA2 | 5q31 | O15460 | details | | Prolyl 4-hydroxylase subunit alpha-1 | P4HA1 | 10q21.3-q23.1 | P13674 | details | | Gamma-butyrobetaine dioxygenase | BBOX1 | 11p14.2 | O75936 | details | | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | PLOD1 | 1p36.22 | Q02809 | details | | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 | PLOD2 | 3q24 | O00469 | details | | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 | PLOD3 | 7q22 | O60568 | details | | Succinyl-CoA:3-ketoacid coenzyme A transferase 1, mitochondrial | OXCT1 | 5p13.1 | P55809 | details | | Succinyl-CoA:3-ketoacid coenzyme A transferase 2, mitochondrial | OXCT2 | 1p34 | Q9BYC2 | details | | Succinyl-CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial | SUCLG1 | 2p11.2 | P53597 | details | | Succinyl-CoA ligase [GDP-forming] subunit beta, mitochondrial | SUCLG2 | 3p14.1 | Q96I99 | details | | Succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial | SUCLA2 | 13q12.2-q13.3 | Q9P2R7 | details | | Aspartyl/asparaginyl beta-hydroxylase | ASPH | 8q12.1 | Q12797 | details | | Mitochondrial dicarboxylate carrier | SLC25A10 | 17q25.3 | Q9UBX3 | details | | 17-beta-hydroxysteroid dehydrogenase type 6 | HSD17B6 | 12q13 | O14756 | details | | Trimethyllysine dioxygenase, mitochondrial | TMLHE | Xq28 | Q9NVH6 | details | | Polycystic kidney disease 2-like 1 protein | PKD2L1 | 10q24 | Q9P0L9 | details | | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2 | ALKBH2 | 12q24.11 | Q6NS38 | details | | Prolyl 3-hydroxylase 1 | LEPRE1 | 1p34.1 | Q32P28 | details | | Prolyl 3-hydroxylase 2 | LEPREL1 | 3q28 | Q8IVL5 | details | | Prolyl 3-hydroxylase 3 | LEPREL2 | 12q13 | Q8IVL6 | details | | Prolyl 4-hydroxylase subunit alpha-3 | P4HA3 | 11q13.4 | Q7Z4N8 | details | | Lysine-specific demethylase 2A | KDM2A | 11q13.2 | Q9Y2K7 | details | | Lysine-specific demethylase 2B | KDM2B | 12q24.31 | Q8NHM5 | details | | Egl nine homolog 1 | EGLN1 | 1q42.1 | Q9GZT9 | details | | Egl nine homolog 2 | EGLN2 | 19q13.2 | Q96KS0 | details | | Egl nine homolog 3 | EGLN3 | 14q13.1 | Q9H6Z9 | details | | Transmembrane prolyl 4-hydroxylase | P4HTM | 3p21.3 | Q9NXG6 | details | | Lysine-specific demethylase 8 | KDM8 | 16p12.1 | Q8N371 | details | | Bifunctional lysine-specific demethylase and histidyl-hydroxylase MINA | MINA | 3q11.2 | Q8IUF8 | details | | Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66 | NO66 | 14q24.3 | Q9H6W3 | details | | Histone lysine demethylase PHF8 | PHF8 | Xp11.22 | Q9UPP1 | details | | Methylcytosine dioxygenase TET1 | TET1 | 10q21 | Q8NFU7 | details | | Methylcytosine dioxygenase TET2 | TET2 | 4q24 | Q6N021 | details | | Methylcytosine dioxygenase TET3 | TET3 | 2p13.1 | O43151 | details |
|
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 0.03368 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 0.0383 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 0.0379 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.0291 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 0.0616 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 0.0289 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000254 |
|---|
| DrugBank ID | DB00139 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB001931 |
|---|
| KNApSAcK ID | C00001205 |
|---|
| Chemspider ID | 1078 |
|---|
| KEGG Compound ID | C00042 |
|---|
| BioCyc ID | SUC |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Succinic_acid |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 1738118 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15741 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
- Thakker C, Martinez I, San KY, Bennett GN: Succinate production in Escherichia coli. Biotechnol J. 2012 Feb;7(2):213-24. doi: 10.1002/biot.201100061. Epub 2011 Sep 20. [PubMed:21932253 ]
- Briere JJ, Favier J, El Ghouzzi V, Djouadi F, Benit P, Gimenez AP, Rustin P: Succinate dehydrogenase deficiency in human. Cell Mol Life Sci. 2005 Oct;62(19-20):2317-24. doi: 10.1007/s00018-005-5237-6. [PubMed:16143825 ]
- Rustin P, Rotig A: Inborn errors of complex II--unusual human mitochondrial diseases. Biochim Biophys Acta. 2002 Jan 17;1553(1-2):117-22. doi: 10.1016/s0005-2728(01)00228-6. [PubMed:11803021 ]
- Yang M, Soga T, Pollard PJ: Oncometabolites: linking altered metabolism with cancer. J Clin Invest. 2013 Sep;123(9):3652-8. doi: 10.1172/JCI67228. Epub 2013 Sep 3. [PubMed:23999438 ]
- Sciacovelli M, Frezza C: Oncometabolites: Unconventional triggers of oncogenic signalling cascades. Free Radic Biol Med. 2016 Nov;100:175-181. doi: 10.1016/j.freeradbiomed.2016.04.025. Epub 2016 Apr 23. [PubMed:27117029 ]
- Gupta A, Dwivedi M, Mahdi AA, Khetrapal CL, Bhandari M: Broad identification of bacterial type in urinary tract infection using (1)h NMR spectroscopy. J Proteome Res. 2012 Mar 2;11(3):1844-54. doi: 10.1021/pr2010692. Epub 2012 Jan 31. [PubMed:22292465 ]
- Sauer M, Porro D, Mattanovich D, Branduardi P: Microbial production of organic acids: expanding the markets. Trends Biotechnol. 2008 Feb;26(2):100-8. doi: 10.1016/j.tibtech.2007.11.006. Epub 2008 Jan 11. [PubMed:18191255 ]
- Becker J, Lange A, Fabarius J, Wittmann C: Top value platform chemicals: bio-based production of organic acids. Curr Opin Biotechnol. 2015 Dec;36:168-75. doi: 10.1016/j.copbio.2015.08.022. Epub 2015 Sep 8. [PubMed:26360870 ]
- Gunnarsson IB, Kuglarz M, Karakashev D, Angelidaki I: Thermochemical pretreatments for enhancing succinic acid production from industrial hemp (Cannabis sativa L.). Bioresour Technol. 2015 Apr;182:58-66. doi: 10.1016/j.biortech.2015.01.126. Epub 2015 Feb 4. [PubMed:25682224 ]
|
|---|


