| Identification |
|---|
| HMDB Protein ID
| CDBP04361 |
| Secondary Accession Numbers
| Not Available |
| Name
| Egl nine homolog 1 |
| Description
| Not Available |
| Synonyms
|
- HIF-PH2
- HIF-prolyl hydroxylase 2
- HPH-2
- Hypoxia-inducible factor prolyl hydroxylase 2
- PHD2
- Prolyl hydroxylase domain-containing protein 2
- SM-20
|
| Gene Name
| EGLN1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in oxidoreductase activity |
| Specific Function
| Cellular oxygen sensor that catalyzes, under normoxic conditions, the post-translational formation of 4-hydroxyproline in hypoxia-inducible factor (HIF) alpha proteins. Hydroxylates a specific proline found in each of the oxygen-dependent degradation (ODD) domains (N-terminal, NODD, and C-terminal, CODD) of HIF1A. Also hydroxylates HIF2A. Has a preference for the CODD site for both HIF1A and HIF1B. Hydroxylated HIFs are then targeted for proteasomal degradation via the von Hippel-Lindau ubiquitination complex. Under hypoxic conditions, the hydroxylation reaction is attenuated allowing HIFs to escape degradation resulting in their translocation to the nucleus, heterodimerization with HIF1B, and increased expression of hypoxy-inducible genes. EGLN1 is the most important isozyme under normoxia and, through regulating the stability of HIF1, involved in various hypoxia-influenced processes such as angiogenesis in retinal and cardiac functionality.
|
| GO Classification
|
| Biological Process |
| labyrinthine layer development |
| negative regulation of sequence-specific DNA binding transcription factor activity |
| oxygen homeostasis |
| peptidyl-proline hydroxylation to 4-hydroxy-L-proline |
| regulation of angiogenesis |
| response to nitric oxide |
| ventricular septum morphogenesis |
| regulation of transcription from RNA polymerase II promoter in response to hypoxia |
| cardiac muscle tissue morphogenesis |
| heart trabecula formation |
| Cellular Component |
| cytosol |
| nucleus |
| Function |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| catalytic activity |
| transition metal ion binding |
| zinc ion binding |
| l-ascorbic acid binding |
| iron ion binding |
| oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| vitamin binding |
| oxidoreductase activity |
| Molecular Function |
| peptidyl-proline 4-dioxygenase activity |
| oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen |
| L-ascorbic acid binding |
| iron ion binding |
| zinc ion binding |
| Process |
| metabolic process |
| oxidation reduction |
|
| Cellular Location
|
Not Available
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | HIF-1 signaling pathway | Not Available | 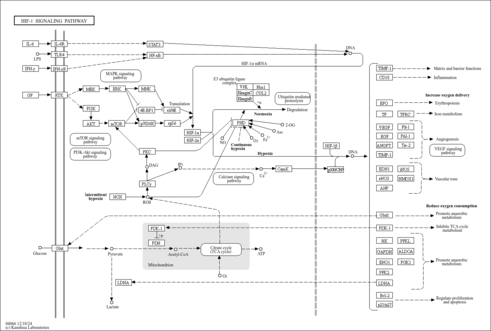 | | Renal cell carcinoma | Not Available | 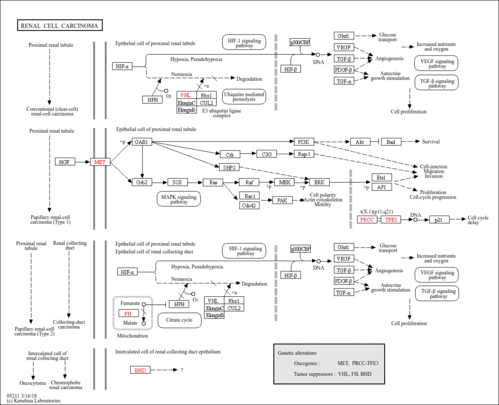 | | The Oncogenic Action of Succinate |    | Not Available | | The Oncogenic Action of Fumarate | 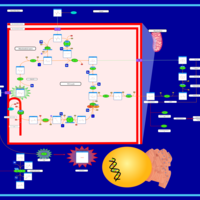 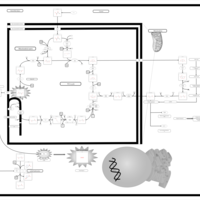  | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 1 |
| Locus
| 1q42.1 |
| SNPs
| EGLN1 |
| Gene Sequence
|
>1281 bp
ATGGCCAATGACAGCGGCGGGCCCGGCGGGCCGAGCCCGAGCGAGCGAGACCGGCAGTAC
TGCGAGCTGTGCGGGAAGATGGAGAACCTGCTGCGCTGCAGCCGCTGCCGCAGCTCCTTC
TACTGCTGCAAGGAGCACCAGCGTCAGGACTGGAAGAAGCACAAGCTCGTGTGCCAGGGC
AGCGAGGGCGCCCTCGGCCACGGAGTGGGCCCACACCAGCATTCCGGCCCCGCGCCGCCG
GCTGCAGTGCCGCCGCCCAGGGCCGGGGCCCGGGAGCCCAGGAAGGCAGCGGCGCGCCGG
GACAACGCCTCCGGGGACGCGGCCAAGGGAAAAGTAAAGGCCAAGCCCCCGGCCGACCCA
GCGGCGGCCGCGTCGCCGTGTCGTGCGGCCGCCGGCGGCCAGGGCTCGGCGGTGGCTGCC
GAAGCCGAGCCCGGCAAGGAGGAGCCGCCGGCCCGCTCATCGCTGTTCCAGGAGAAGGCG
AACCTGTACCCCCCAAGCAACACGCCCGGGGATGCGCTGAGCCCCGGCGGCGGCCTGCGG
CCCAACGGGCAGACGAAGCCCCTGCCGGCGCTGAAGCTGGCGCTCGAGTACATCGTGCCG
TGCATGAACAAGCACGGCATCTGTGTGGTGGACGACTTCCTCGGCAAGGAGACCGGACAG
CAGATCGGCGACGAGGTGCGCGCCCTGCACGACACCGGGAAGTTCACGGACGGGCAGCTG
GTCAGCCAGAAGAGTGACTCGTCCAAGGACATCCGAGGCGATAAGATCACCTGGATCGAG
GGCAAGGAGCCCGGCTGCGAAACCATTGGGCTGCTCATGAGCAGCATGGACGACCTGATA
CGCCACTGTAACGGGAAGCTGGGCAGCTACAAAATCAATGGCCGGACGAAAGCCATGGTT
GCTTGTTATCCGGGCAATGGAACGGGTTATGTACGTCATGTTGATAATCCAAATGGAGAT
GGAAGATGTGTGACATGTATATATTATCTTAATAAAGACTGGGATGCCAAGGTAAGTGGA
GGTATACTTCGAATTTTTCCAGAAGGCAAAGCCCAGTTTGCTGACATTGAACCCAAATTT
GATAGACTGCTGTTTTTCTGGTCTGACCGTCGCAACCCTCATGAAGTACAACCAGCATAT
GCTACAAGGTACGCAATAACTGTTTGGTATTTTGATGCAGATGAGAGAGCACGAGCTAAA
GTAAAATATCTAACAGGTGAAAAAGGTGTGAGGGTTGAACTCAATAAACCTTCAGATTCG
GTCGGTAAAGACGTCTTCTAG
|
| Protein Properties |
|---|
| Number of Residues
| 426 |
| Molecular Weight
| 46020.585 |
| Theoretical pI
| 8.534 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Egl nine homolog 1
MANDSGGPGGPSPSERDRQYCELCGKMENLLRCSRCRSSFYCCKEHQRQDWKKHKLVCQG
SEGALGHGVGPHQHSGPAPPAAVPPPRAGAREPRKAAARRDNASGDAAKGKVKAKPPADP
AAAASPCRAAAGGQGSAVAAEAEPGKEEPPARSSLFQEKANLYPPSNTPGDALSPGGGLR
PNGQTKPLPALKLALEYIVPCMNKHGICVVDDFLGKETGQQIGDEVRALHDTGKFTDGQL
VSQKSDSSKDIRGDKITWIEGKEPGCETIGLLMSSMDDLIRHCNGKLGSYKINGRTKAMV
ACYPGNGTGYVRHVDNPNGDGRCVTCIYYLNKDWDAKVSGGILRIFPEGKAQFADIEPKF
DRLLFFWSDRRNPHEVQPAYATRYAITVWYFDADERARAKVKYLTGEKGVRVELNKPSDS
VGKDVF
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| Q9GZT9 |
| UniProtKB/Swiss-Prot Entry Name
| EGLN1_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AF229245 |
| GeneCard ID
| EGLN1 |
| GenAtlas ID
| EGLN1 |
| HGNC ID
| HGNC:1232 |
| References |
|---|
| General References
| Not Available |

