| Identification |
|---|
| HMDB Protein ID
| CDBP04363 |
| Secondary Accession Numbers
| Not Available |
| Name
| Egl nine homolog 3 |
| Description
| Not Available |
| Synonyms
|
- HIF-PH3
- HIF-prolyl hydroxylase 3
- HPH-1
- HPH-3
- Hypoxia-inducible factor prolyl hydroxylase 3
- PHD3
- Prolyl hydroxylase domain-containing protein 3
|
| Gene Name
| EGLN3 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in oxidoreductase activity |
| Specific Function
| Cellular oxygen sensor that catalyzes, under normoxic conditions, the post-translational formation of 4-hydroxyproline in hypoxia-inducible factor (HIF) alpha proteins. Hydroxylates a specific proline found in each of the oxygen-dependent degradation (ODD) domains (N-terminal, NODD, and C-terminal, CODD) of HIF1A. Also hydroxylates HIF2A. Has a preference for the CODD site for both HIF1A and HIF2A. Hydroxylation on the NODD site by EGLN3 appears to require prior hydroxylation on the CODD site. Hydroxylated HIFs are then targeted for proteasomal degradation via the von Hippel-Lindau ubiquitination complex. Under hypoxic conditions, the hydroxylation reaction is attenuated allowing HIFs to escape degradation resulting in their translocation to the nucleus, heterodimerization with HIF1B, and increased expression of hypoxy-inducible genes. EGLN3 is the most important isozyme in limiting physiological activation of HIFs (particularly HIF2A) in hypoxia. Also hydroxylates PKM in hypoxia, limiting glycolysis. Under normoxia, hydroxylates and regulates the stability of ADRB2. Regulator of cardiomyocyte and neuronal apoptosis. In cardiomyocytes, inhibits the anti-apoptotic effect of BCL2 by disrupting the BAX-BCL2 complex. In neurons, has a NGF-induced proapoptotic effect, probably through regulating CASP3 activity. Also essential for hypoxic regulation of neutrophilic inflammation. Plays a crucial role in DNA damage response (DDR) by hydroxylating TELO2, promoting its interaction with ATR which is required for activation of the ATR/CHK1/p53 pathway.
|
| GO Classification
|
| Biological Process |
| regulation of cell proliferation |
| activation of cysteine-type endopeptidase activity involved in apoptotic process |
| response to DNA damage stimulus |
| regulation of transcription from RNA polymerase II promoter in response to hypoxia |
| peptidyl-proline hydroxylation to 4-hydroxy-L-proline |
| regulation of neuron apoptotic process |
| protein hydroxylation |
| Cellular Component |
| cytosol |
| nucleoplasm |
| Function |
| oxidoreductase activity |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| catalytic activity |
| transition metal ion binding |
| l-ascorbic acid binding |
| iron ion binding |
| oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| vitamin binding |
| Molecular Function |
| oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen |
| L-ascorbic acid binding |
| iron ion binding |
| peptidyl-proline 4-dioxygenase activity |
| Process |
| metabolic process |
| oxidation reduction |
|
| Cellular Location
|
- Nucleus
- Cytoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | HIF-1 signaling pathway | Not Available | 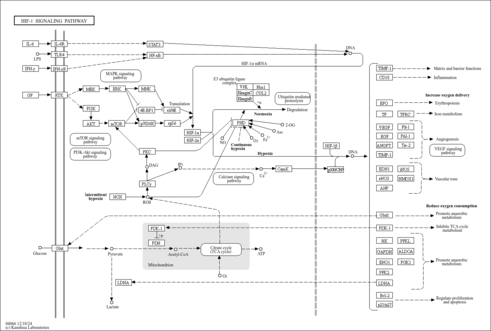 | | Renal cell carcinoma | Not Available | 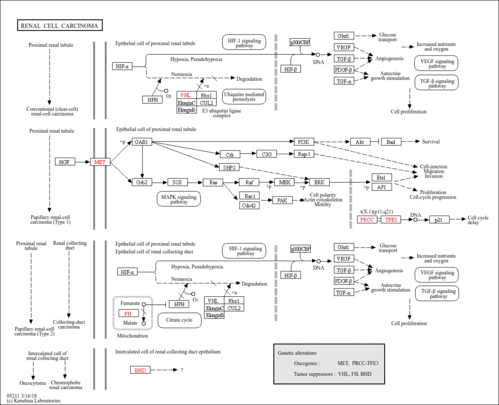 | | The Oncogenic Action of Succinate |    | Not Available | | The Oncogenic Action of Fumarate | 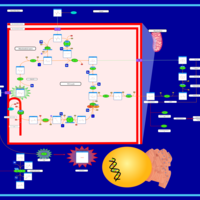 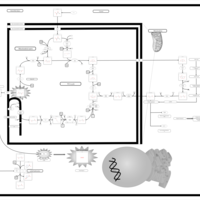  | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 14 |
| Locus
| 14q13.1 |
| SNPs
| EGLN3 |
| Gene Sequence
|
>720 bp
ATGCCCCTGGGACACATCATGAGGCTGGACCTGGAGAAAATTGCCCTGGAGTACATCGTG
CCCTGTCTGCACGAGGTGGGCTTCTGCTACCTGGACAACTTCCTGGGCGAGGTGGTGGGC
GACTGCGTCCTGGAGCGCGTCAAGCAGCTGCACTGCACCGGGGCCCTGCGGGACGGCCAG
CTGGCGGGGCCGCGCGCCGGCGTCTCCAAGCGACACCTGCGGGGCGACCAGATCACGTGG
ATCGGGGGCAACGAGGAGGGCTGCGAGGCCATCAGCTTCCTCCTGTCCCTCATCGACAGG
CTGGTCCTCTACTGCGGGAGCCGGCTGGGCAAATACTACGTCAAGGAGAGGTCTAAGGCA
ATGGTGGCTTGCTATCCGGGAAATGGAACAGGTTATGTTCGCCACGTGGACAACCCCAAC
GGTGATGGTCGCTGCATCACCTGCATCTACTATCTGAACAAGAATTGGGATGCCAAGCTA
CATGGTGGGATCCTGCGGATATTTCCAGAGGGGAAATCATTCATAGCAGATGTGGAGCCC
ATTTTTGACAGACTCCTGTTCTTCTGGTCAGATCGTAGGAACCCACACGAAGTGCAGCCC
TCTTACGCAACCAGATATGCTATGACTGTCTGGTACTTTGATGCTGAAGAAAGGGCAGAA
GCCAAAAAGAAATTCAGGAATTTAACTAGGAAAACTGAATCTGCCCTCACTGAAGACTGA
|
| Protein Properties |
|---|
| Number of Residues
| 239 |
| Molecular Weight
| 27261.06 |
| Theoretical pI
| 7.636 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Egl nine homolog 3
MPLGHIMRLDLEKIALEYIVPCLHEVGFCYLDNFLGEVVGDCVLERVKQLHCTGALRDGQ
LAGPRAGVSKRHLRGDQITWIGGNEEGCEAISFLLSLIDRLVLYCGSRLGKYYVKERSKA
MVACYPGNGTGYVRHVDNPNGDGRCITCIYYLNKNWDAKLHGGILRIFPEGKSFIADVEP
IFDRLLFFWSDRRNPHEVQPSYATRYAMTVWYFDAEERAEAKKKFRNLTRKTESALTED
|
| External Links |
|---|
| GenBank ID Protein
| 14547150 |
| UniProtKB/Swiss-Prot ID
| Q9H6Z9 |
| UniProtKB/Swiss-Prot Entry Name
| EGLN3_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| AJ310545 |
| GeneCard ID
| EGLN3 |
| GenAtlas ID
| EGLN3 |
| HGNC ID
| HGNC:14661 |
| References |
|---|
| General References
| Not Available |

