| Identification |
|---|
| HMDB Protein ID
| CDBP04362 |
| Secondary Accession Numbers
| Not Available |
| Name
| Egl nine homolog 2 |
| Description
| Not Available |
| Synonyms
|
- Estrogen-induced tag 6
- HIF-PH1
- HIF-prolyl hydroxylase 1
- HPH-1
- HPH-3
- Hypoxia-inducible factor prolyl hydroxylase 1
- PHD1
- Prolyl hydroxylase domain-containing protein 1
|
| Gene Name
| EGLN2 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in oxidoreductase activity |
| Specific Function
| Cellular oxygen sensor that catalyzes, under normoxic conditions, the post-translational formation of 4-hydroxyproline in hypoxia-inducible factor (HIF) alpha proteins. Hydroxylates a specific proline found in each of the oxygen-dependent degradation (ODD) domains (N-terminal, NODD, and C-terminal, CODD) of HIF1A. Also hydroxylates HIF2A. Has a preference for the CODD site for both HIF1A and HIF2A. Hydroxylated HIFs are then targeted for proteasomal degradation via the von Hippel-Lindau ubiquitination complex. Under hypoxic conditions, the hydroxylation reaction is attenuated allowing HIFs to escape degradation resulting in their translocation to the nucleus, heterodimerization with HIF1B, and increased expression of hypoxy-inducible genes. EGLN2 is involved in regulating hypoxia tolerance and apoptosis in cardiac and skeletal muscle. Also regulates susceptibility to normoxic oxidative neuronal death.
|
| GO Classification
|
| Biological Process |
| cell redox homeostasis |
| peptidyl-proline hydroxylation to 4-hydroxy-L-proline |
| regulation of cell growth |
| positive regulation of protein catabolic process |
| regulation of neuron apoptotic process |
| intracellular estrogen receptor signaling pathway |
| regulation of transcription from RNA polymerase II promoter in response to hypoxia |
| Cellular Component |
| nucleoplasm |
| Function |
| l-ascorbic acid binding |
| iron ion binding |
| oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen |
| vitamin binding |
| oxidoreductase activity |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| catalytic activity |
| transition metal ion binding |
| Molecular Function |
| oxygen sensor activity |
| ferrous iron binding |
| oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen |
| L-ascorbic acid binding |
| peptidyl-proline 4-dioxygenase activity |
| Process |
| metabolic process |
| oxidation reduction |
|
| Cellular Location
|
- Nucleus
- Cytoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | HIF-1 signaling pathway | Not Available | 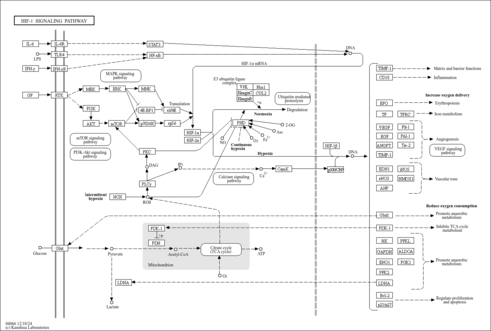 | | Renal cell carcinoma | Not Available | 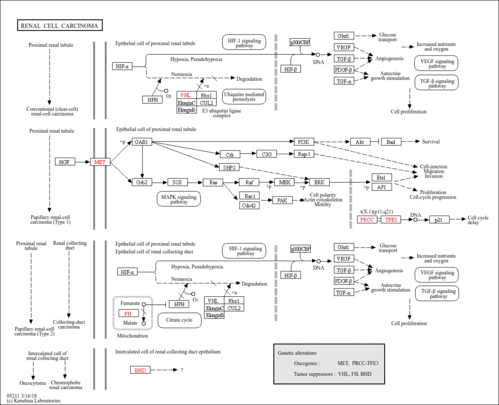 | | The Oncogenic Action of Succinate |    | Not Available | | The Oncogenic Action of Fumarate | 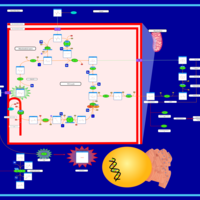 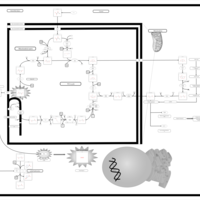  | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 19 |
| Locus
| 19q13.2 |
| SNPs
| EGLN2 |
| Gene Sequence
|
>1224 bp
ATGGACAGCCCGTGCCAGCCGCAGCCCCTAAGTCAGGCTCTCCCTCAGTTACCAGGGTCT
TCGTCAGAGCCCTTGGAGCCTGAGCCTGGCCGGGCCAGGATGGGAGTGGAGAGTTACCTG
CCCTGTCCCCTGCTCCCCTCCTACCACTGTCCAGGAGTGCCTAGTGAGGCCTCGGCAGGG
AGTGGGACCCCCAGAGCCACAGCCACCTCTACCACTGCCAGCCCTCTTCGGGACGGTTTT
GGCGGGCAGGATGGTGGTGAGCTGCGGCCGCTGCAGAGTGAAGGCGCTGCAGCGCTGGTC
ACCAAGGGGTGCCAGCGATTGGCAGCCCAGGGCGCACGGCCTGAGGCCCCCAAACGGAAA
TGGGCCGAGGATGGTGGGGATGCCCCTTCACCCAGCAAACGGCCCTGGGCCAGGCAAGAG
AACCAGGAGGCAGAGCGGGAGGGTGGCATGAGCTGCAGCTGCAGCAGTGGCAGTGGTGAG
GCCAGTGCTGGGCTGATGGAGGAGGCGCTGCCCTCTGCGCCCGAGCGCCTGGCCCTGGAC
TATATCGTGCCCTGCATGCGGTACTACGGCATCTGCGTCAAGGACAGCTTCCTGGGGGCA
GCACTGGGCGGTCGCGTGCTGGCCGAGGTGGAGGCCCTCAAACGGGGTGGGCGCCTGCGA
GACGGGCAGCTAGTGAGCCAGAGGGCGATCCCGCCGCGCAGCATCCGTGGGGACCAGATT
GCCTGGGTGGAAGGCCATGAACCAGGCTGTCGAAGCATTGGTGCCCTCATGGCCCATGTG
GACGCCGTCATCCGCCACTGCGCAGGGCGGCTGGGCAGCTATGTCATCAACGGGCGCACC
AAGGCCATGGTGGCGTGTTACCCAGGCAACGGGCTCGGGTACGTAAGGCACGTTGACAAT
CCCCACGGCGATGGGCGCTGCATCACCTGTATCTATTACCTGAATCAGAACTGGGACGTT
AAGGTGCATGGCGGCCTGCTGCAGATCTTCCCTGAGGGCCGGCCCGTGGTAGCCAACATC
GAGCCACTCTTTGACCGGTTGCTCATTTTCTGGTCTGACCGGCGGAACCCCCACGAGGTG
AAGCCAGCCTATGCCACCAGGTACGCCATCACTGTCTGGTATTTTGATGCCAAGGAGCGG
GCAGCAGCCAAAGACAAGTATCAGCTAGCATCAGGACAGAAAGGTGTCCAAGTACCTGTA
TCACAGCCGCCTACGCCCACCTAG
|
| Protein Properties |
|---|
| Number of Residues
| 407 |
| Molecular Weight
| 43650.03 |
| Theoretical pI
| 7.908 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Egl nine homolog 2
MDSPCQPQPLSQALPQLPGSSSEPLEPEPGRARMGVESYLPCPLLPSYHCPGVPSEASAG
SGTPRATATSTTASPLRDGFGGQDGGELRPLQSEGAAALVTKGCQRLAAQGARPEAPKRK
WAEDGGDAPSPSKRPWARQENQEAEREGGMSCSCSSGSGEASAGLMEEALPSAPERLALD
YIVPCMRYYGICVKDSFLGAALGGRVLAEVEALKRGGRLRDGQLVSQRAIPPRSIRGDQI
AWVEGHEPGCRSIGALMAHVDAVIRHCAGRLGSYVINGRTKAMVACYPGNGLGYVRHVDN
PHGDGRCITCIYYLNQNWDVKVHGGLLQIFPEGRPVVANIEPLFDRLLIFWSDRRNPHEV
KPAYATRYAITVWYFDAKERAAAKDKYQLASGQKGVQVPVSQPPTPT
|
| External Links |
|---|
| GenBank ID Protein
| 14547148 |
| UniProtKB/Swiss-Prot ID
| Q96KS0 |
| UniProtKB/Swiss-Prot Entry Name
| EGLN2_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| AJ310544 |
| GeneCard ID
| EGLN2 |
| GenAtlas ID
| EGLN2 |
| HGNC ID
| HGNC:14660 |
| References |
|---|
| General References
| Not Available |

