| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:27:40 UTC |
|---|
| Updated at | 2020-12-07 19:07:13 UTC |
|---|
| CannabisDB ID | CDB000178 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | alpha-Linolenic acid |
|---|
| Description | alpha-Linolenic acid (ALA) is a polyunsaturated fatty acid (PUFA). It is a member of the group of essential fatty acids called omega-3 fatty acids. alpha-Linolenic acid is not synthesized by mammals and therefore is an essential dietary requirement for all mammals. Omega-3 fatty acids get their name based on the location of one of their first double bonds. In all omega-3 fatty acids, the first double bond is located between the third and fourth carbon atom counting from the methyl end of the fatty acid (n-3). Although humans and other mammals can synthesize saturated and some monounsaturated fatty acids from carbon groups in carbohydrates and proteins, they lack the enzymes necessary to insert a cis double bond at the n-6 or the n-3 position of a fatty acid. Certain nuts (English walnuts) and vegetable oils (canola, soybean, flaxseed/linseed, olive) are rich in alpha-linolenic acid. Linolenic acid is a fatty acid found in hemp seed oil of cannabis plants, first reported by Prior et al. in 1968 (PMID: 6991645 ). Omega-3 fatty acids like alpha-linolenic acid are important structural components of cell membranes. When incorporated into phospholipids, they affect cell membrane properties such as fluidity, flexibility, permeability, and the activity of membrane-bound enzymes. Omega-3 fatty acids can modulate the expression of several genes, including those involved with fatty acid metabolism and inflammation. alpha-Linolenic acid and other omega-3 fatty acids may regulate gene expression by interacting with specific transcription factors, including peroxisome proliferator-activated receptors (PPARs) and liver X receptors (LXRs). alpha-Linolenic acid is found to be associated with isovaleric acidemia, which is an inborn error of metabolism. Patients with isovaleric acidemia have a defect in leucine catabolism resulting from a deficiency in the enzyme isovaleryl-CoA dehydrogenase. These patients do well on a low protein/low leucine diet but a low protein diet can lead to deficiencies in essential fatty acids, such as alpha-linolenic acid ( Ref:DOI ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (9,12,15)-Linolenic acid | ChEBI | | (9Z,12Z,15Z)-Octadecatrienoic acid | ChEBI | | (Z,Z,Z)-9,12,15-Octadecatrienoic acid | ChEBI | | 9,12,15-Octadecatrienoic acid | ChEBI | | 9-cis,12-cis,15-cis-Octadecatrienoic acid | ChEBI | | ALA | ChEBI | | all-cis-9,12,15-Octadecatrienoic acid | ChEBI | | cis,cis,cis-9,12,15-Octadecatrienoic acid | ChEBI | | cis-Delta(9,12,15)-Octadecatrienoic acid | ChEBI | | Linolenic acid | ChEBI | | Linolenate | Kegg | | alpha-Linolenate | Kegg | | (9,12,15)-Linolenate | Generator | | (9Z,12Z,15Z)-Octadecatrienoate | Generator | | (Z,Z,Z)-9,12,15-Octadecatrienoate | Generator | | 9,12,15-Octadecatrienoate | Generator | | 9-cis,12-cis,15-cis-Octadecatrienoate | Generator | | all-cis-9,12,15-Octadecatrienoate | Generator | | cis,cis,cis-9,12,15-Octadecatrienoate | Generator | | cis-delta(9,12,15)-Octadecatrienoate | Generator | | cis-Δ(9,12,15)-octadecatrienoate | Generator | | cis-Δ(9,12,15)-octadecatrienoic acid | Generator | | a-Linolenate | Generator | | a-Linolenic acid | Generator | | Α-linolenate | Generator | | Α-linolenic acid | Generator | | alpha Linolenic acid | MeSH | | cis-9,12,15-Octadecatrienoate | HMDB | | cis-9,12,15-Octadecatrienoic acid | HMDB | | Industrene 120 | HMDB | | FA(18:3(9Z,12Z,15Z)) | HMDB | | FA(18:3n3) | HMDB |
|
|---|
| Chemical Formula | C18H30O2 |
|---|
| Average Molecular Weight | 278.43 |
|---|
| Monoisotopic Molecular Weight | 278.2246 |
|---|
| IUPAC Name | (9Z,12Z,15Z)-octadeca-9,12,15-trienoic acid |
|---|
| Traditional Name | α linolenic acid |
|---|
| CAS Registry Number | 463-40-1 |
|---|
| SMILES | CC\C=C/C\C=C/C\C=C/CCCCCCCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C18H30O2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18(19)20/h3-4,6-7,9-10H,2,5,8,11-17H2,1H3,(H,19,20)/b4-3-,7-6-,10-9- |
|---|
| InChI Key | DTOSIQBPPRVQHS-PDBXOOCHSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as lineolic acids and derivatives. These are derivatives of lineolic acid. Lineolic acid is a polyunsaturated omega-6 18 carbon long fatty acid, with two CC double bonds at the 9- and 12-positions. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Lineolic acids and derivatives |
|---|
| Direct Parent | Lineolic acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Octadecanoid
- Long-chain fatty acid
- Fatty acid
- Unsaturated fatty acid
- Straight chain fatty acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | -16.5 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | 6.46 | SANGSTER (1993) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | alpha-Linolenic acid, 1 TMS, GC-MS Spectrum | splash10-052f-7900000000-8765cf82603feb1b448f | Spectrum | | GC-MS | alpha-Linolenic acid, 1 TMS, GC-MS Spectrum | splash10-004i-9300000000-302519a616eaed5fcc59 | Spectrum | | GC-MS | alpha-Linolenic acid, 1 TMS, GC-MS Spectrum | splash10-005c-9800000000-2d44b3c70e2992b9a812 | Spectrum | | GC-MS | alpha-Linolenic acid, non-derivatized, GC-MS Spectrum | splash10-052f-7900000000-8765cf82603feb1b448f | Spectrum | | GC-MS | alpha-Linolenic acid, non-derivatized, GC-MS Spectrum | splash10-004i-9300000000-302519a616eaed5fcc59 | Spectrum | | GC-MS | alpha-Linolenic acid, non-derivatized, GC-MS Spectrum | splash10-005c-9800000000-2d44b3c70e2992b9a812 | Spectrum | | GC-MS | alpha-Linolenic acid, non-derivatized, GC-MS Spectrum | splash10-052f-5900000000-28710ea35f196c595e03 | Spectrum | | Predicted GC-MS | alpha-Linolenic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-052e-6950000000-27037263e74c8119ead0 | Spectrum | | Predicted GC-MS | alpha-Linolenic acid, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00ds-6931000000-657fa0cc66a206a36409 | Spectrum | | Predicted GC-MS | alpha-Linolenic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, N/A (Annotated) | splash10-004i-0190000000-aee97e9ea2c7f0783adf | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, N/A (Annotated) | splash10-05nb-9500000000-fb92aefc701027239359 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, N/A (Annotated) | splash10-0aru-9100000000-a5dddfe6f629d1371fac | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - ESI-TOF 10V, Negative | splash10-004i-0090000000-2b7bea03a685454dd135 | 2017-09-12 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - ESI-TOF , Negative | splash10-004i-0090000000-53ddfaed6ce13c37d957 | 2017-09-12 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - ESI-TOF 20V, Negative | splash10-004i-0090000000-e5350a66361bc63d1071 | 2017-09-12 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - ESI-TOF 30V, Negative | splash10-004i-0090000000-5731aa5e301022c06813 | 2017-09-12 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - ESI-TOF 10V, Negative | splash10-004i-0091010000-6922411e48d747e592b5 | 2017-09-12 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , negative | splash10-001i-0090000000-a18b573ad888d048efa6 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-TOF , negative | splash10-004i-0090000000-2b7bea03a685454dd135 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-TOF , negative | splash10-004i-0090000000-e5350a66361bc63d1071 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-TOF , negative | splash10-004i-0090000000-5731aa5e301022c06813 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 10V, positive | splash10-004i-0090000000-1d391027db3704d71cb9 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 20V, positive | splash10-00lr-9210000000-41acd8a55f52afec6d79 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 40V, positive | splash10-014i-9000000000-a976b4a80a07eb2c4641 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9100000000-497eddf5d738a45f83fb | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0090000000-0c00ca98901526cb2e26 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-004i-1090000000-35ffce9343fe0d15ea0d | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-004i-0090000000-373584fbab950d6676cc | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01t9-0090000000-ae1f482177d0e208165e | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00o0-5690000000-20804c8382f14ff0ccf4 | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014l-8930000000-9d5e342b1351adc71c20 | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0090000000-4d3e8d1180800a7b4ed5 | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0059-1090000000-67ab14c068a142dd3c30 | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9230000000-403c610d63380e63109a | 2017-07-26 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, CDCl3, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Alpha Linolenic Acid and Linoleic Acid Metabolism |    | 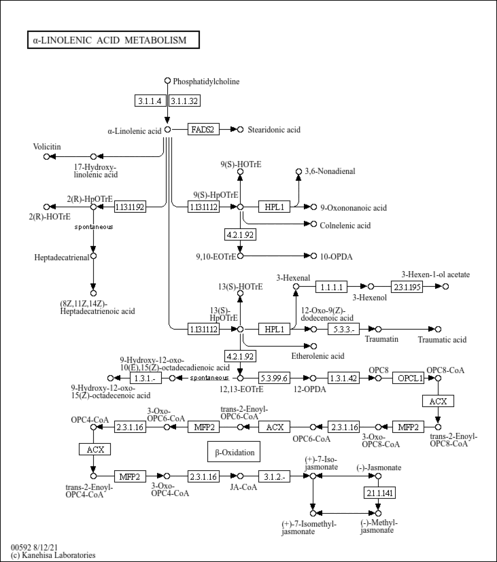 |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
| Transient receptor potential cation channel subfamily V member 1 | TRPV1 | 17p13.2 | Q8NER1 | details | | Sodium/calcium exchanger 1 | SLC8A1 | 2p23-p22 | P32418 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | |
| Phospholipase A2 | PLA2G1B | 12q23-q24.1 | P04054 | details | | Peroxisome proliferator-activated receptor gamma | PPARG | 3p25 | P37231 | details | | Peroxisome proliferator-activated receptor alpha | PPARA | 22q12-q13.1|22q13.31 | Q07869 | details | | Transient receptor potential cation channel subfamily V member 1 | TRPV1 | 17p13.2 | Q8NER1 | details | | Peroxisome proliferator-activated receptor delta | PPARD | 6p21.2 | Q03181 | details | | Elongation of very long chain fatty acids protein 4 | ELOVL4 | 6q14 | Q9GZR5 | details | | Oxysterols receptor LXR-beta | NR1H2 | 19q13.33 | P55055 | details | | Oxysterols receptor LXR-alpha | NR1H3 | 11p11.2 | Q13133 | details |
|
|---|
| Transcriptional Factors | |
| Phospholipase A2 | PLA2G1B | 12q23-q24.1 | P04054 | details | | Group 10 secretory phospholipase A2 | PLA2G10 | 16p13.1-p12 | O15496 | details | | Peroxisome proliferator-activated receptor gamma | PPARG | 3p25 | P37231 | details | | Peroxisome proliferator-activated receptor alpha | PPARA | 22q12-q13.1|22q13.31 | Q07869 | details | | Peroxisome proliferator-activated receptor delta | PPARD | 6p21.2 | Q03181 | details | | Oxysterols receptor LXR-beta | NR1H2 | 19q13.33 | P55055 | details | | Oxysterols receptor LXR-alpha | NR1H3 | 11p11.2 | Q13133 | details |
|
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0001388 |
|---|
| DrugBank ID | DB00132 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB012462 |
|---|
| KNApSAcK ID | C00007247 |
|---|
| Chemspider ID | 4444437 |
|---|
| KEGG Compound ID | C06427 |
|---|
| BioCyc ID | LINOLENIC_ACID |
|---|
| BiGG ID | 48237 |
|---|
| Wikipedia Link | Alpha-Linolenic_acid |
|---|
| METLIN ID | 6208 |
|---|
| PubChem Compound | 5280934 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 27432 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
|
|---|
