| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:21:26 UTC |
|---|
| Updated at | 2022-12-13 23:36:22 UTC |
|---|
| CannabisDB ID | CDB006139 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Glycine |
|---|
| Description | Glycine, also known as Gly or aminoacetic acid, belongs to the class of organic compounds known as alpha amino acids. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon). Glycine is a very strong basic compound (based on its pKa). Glycine exists in all living species, ranging from bacteria to humans. Glycine is an odorless tasting compound. Outside of the human body, Glycine is found, on average, in the highest concentration within a few different foods, such as gelatins, gelatin desserts, and beluga whales and in a lower concentration in hedge mustards, pumpkinseed sunfish, and prunus (cherry, plum). Glycine has also been detected, but not quantified in, several different foods, such as boysenberries, fruit juices, giant butterburs, ginsengs, and Chinese mustards. Glycine is most important and simple, nonessential amino acid in humans, animals, and many mammals. Generally, glycine is synthesized from choline, serine, hydroxyproline, and threonine through interorgan metabolism in which kidneys and liver are the primarily involved. Glycine acts as precursor for several key metabolites of low molecular weight such as creatine, glutathione, haem, purines, and porphyrins. There are a number of reports supporting the role of supplementary glycine in prevention of many diseases and disorders including cancer. Dietary supplementation of glycine is effectual has been shown to be effective in treating metabolic disorders in patients with cardiovascular diseases ( PMID: 25332814 ), several inflammatory diseases, obesity, cancers, and diabetes ( PMID: 27292783 ). Glycine has also been shown to enhance the quality of sleep and neurological functions. (PMID: 28337245 ) |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Aminoacetic acid | ChEBI | | Aminoessigsaeure | ChEBI | | Aminoethanoic acid | ChEBI | | G | ChEBI | | Gly | ChEBI | | Glycin | ChEBI | | Glycocoll | ChEBI | | Glykokoll | ChEBI | | Glyzin | ChEBI | | H2N-CH2-COOH | ChEBI | | Hgly | ChEBI | | Leimzucker | ChEBI | | Aminoacetate | Generator | | Aminoethanoate | Generator | | 2-Aminoacetate | HMDB | | 2-Aminoacetic acid | HMDB | | Aciport | HMDB | | Amino-acetate | HMDB | | Amino-acetic acid | HMDB | | Glicoamin | HMDB | | Glycolixir | HMDB | | Glycosthene | HMDB | | Gyn-hydralin | HMDB | | Padil | HMDB | | Glycine carbonate (1:1), monosodium salt | HMDB | | Glycine carbonate (2:1), monopotassium salt | HMDB | | Glycine sulfate (3:1) | HMDB | | Glycine, monoammonium salt | HMDB | | Glycine, monosodium salt | HMDB | | Glycine, sodium hydrogen carbonate | HMDB | | Monoammonium salt glycine | HMDB | | Calcium salt glycine | HMDB | | Glycine hydrochloride (2:1) | HMDB | | Glycine phosphate (1:1) | HMDB | | Glycine, monopotasssium salt | HMDB | | Monopotasssium salt glycine | HMDB | | Monosodium salt glycine | HMDB | | Glycine carbonate (2:1), monolithium salt | HMDB | | Glycine carbonate (2:1), monosodium salt | HMDB | | Glycine hydrochloride | HMDB | | Glycine, copper salt | HMDB | | Hydrochloride, glycine | HMDB | | Salt glycine, monoammonium | HMDB | | Acid, aminoacetic | HMDB | | Cobalt salt glycine | HMDB | | Copper salt glycine | HMDB | | Glycine phosphate | HMDB | | Glycine, calcium salt | HMDB | | Glycine, calcium salt (2:1) | HMDB | | Glycine, cobalt salt | HMDB | | Phosphate, glycine | HMDB | | Salt glycine, monosodium | HMDB |
|
|---|
| Chemical Formula | C2H5NO2 |
|---|
| Average Molecular Weight | 75.07 |
|---|
| Monoisotopic Molecular Weight | 75.032 |
|---|
| IUPAC Name | 2-aminoacetic acid |
|---|
| Traditional Name | glycine |
|---|
| CAS Registry Number | 56-40-6 |
|---|
| SMILES | NCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C2H5NO2/c3-1-2(4)5/h1,3H2,(H,4,5) |
|---|
| InChI Key | DHMQDGOQFOQNFH-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as alpha amino acids. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon). |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Alpha amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-amino acid
- Amino acid
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic nitrogen compound
- Organic oxide
- Hydrocarbon derivative
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Primary aliphatic amine
- Organic oxygen compound
- Carbonyl group
- Amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Biological role: Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 262.2 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 249 mg/mL | Not Available | | logP | -3.21 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-001i-9000000000-222d6c3a1ba6afcd7ea9 | 2014-09-20 | View Spectrum | | GC-MS | Glycine, 3 TMS, GC-MS Spectrum | splash10-00dj-2900000000-0ef96bcf06ce475afcdd | Spectrum | | GC-MS | Glycine, non-derivatized, GC-MS Spectrum | splash10-00dj-1900000000-1d289099ac79cfb8bb19 | Spectrum | | GC-MS | Glycine, 3 TMS, GC-MS Spectrum | splash10-00di-7910000000-6c972a683dfb75b69331 | Spectrum | | GC-MS | Glycine, 2 TMS, GC-MS Spectrum | splash10-0udi-0900000000-ef69e38ee6cebc2ece00 | Spectrum | | GC-MS | Glycine, 3 TMS, GC-MS Spectrum | splash10-00di-2910000000-3215b9e40f20c7b306cd | Spectrum | | GC-MS | Glycine, non-derivatized, GC-MS Spectrum | splash10-001i-9000000000-719b7f248956f13a312d | Spectrum | | GC-MS | Glycine, non-derivatized, GC-MS Spectrum | splash10-0udi-0900000000-99b4fc43740b21edc786 | Spectrum | | GC-MS | Glycine, non-derivatized, GC-MS Spectrum | splash10-00di-1910000000-4cff4d14c73acff9442f | Spectrum | | GC-MS | Glycine, non-derivatized, GC-MS Spectrum | splash10-00dj-2900000000-0ef96bcf06ce475afcdd | Spectrum | | GC-MS | Glycine, non-derivatized, GC-MS Spectrum | splash10-00dj-1900000000-1d289099ac79cfb8bb19 | Spectrum | | GC-MS | Glycine, non-derivatized, GC-MS Spectrum | splash10-0002-4960000000-2c6fa028e985c6019854 | Spectrum | | GC-MS | Glycine, non-derivatized, GC-MS Spectrum | splash10-00di-7910000000-6c972a683dfb75b69331 | Spectrum | | GC-MS | Glycine, non-derivatized, GC-MS Spectrum | splash10-00di-2910000000-3215b9e40f20c7b306cd | Spectrum | | GC-MS | Glycine, non-derivatized, GC-MS Spectrum | splash10-0udi-0900000000-ef69e38ee6cebc2ece00 | Spectrum | | GC-MS | Glycine, non-derivatized, GC-MS Spectrum | splash10-00ds-2900000000-ffffed9c78c16a884e4a | Spectrum | | GC-MS | Glycine, non-derivatized, GC-MS Spectrum | splash10-004r-3900000000-f288b50b7b6890429811 | Spectrum | | GC-MS | Glycine, non-derivatized, GC-MS Spectrum | splash10-0udi-1900000000-c76140c31c1f120e4b9d | Spectrum | | Predicted GC-MS | Glycine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-001i-9000000000-f0a2cfbefb9fcd9b6c3e | Spectrum | | Predicted GC-MS | Glycine, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00di-9300000000-b8bbfc1276d5adb1ba04 | Spectrum | | Predicted GC-MS | Glycine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Glycine, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Glycine, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Glycine, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-003r-9000000000-725357e461c898a7451e | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001i-9000000000-9f3930e66b117ad91dca | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-001i-9000000000-b3336097dddbb5e22871 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI RMU-6M) , Positive | splash10-001i-9000000000-719b7f248956f13a312d | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-00di-9000000000-6001578fc511ba3fefef | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-00di-9000000000-79b2a0a9d93de6a62358 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-00di-9000000000-9290dbe208c4744f4431 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-004i-9000000000-342ab462db0835abb3d2 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-0ar1-9010000000-9daadc1d169a8530926d | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-07y0-9220000000-8c7785f1f3aa8052679f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-0ula-9110000000-43ada06fe1b56b4e9fcc | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-017i-9000000000-fbd78fbb48f082235f42 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - CE-ESI-TOF (CE-system connected to 6210 Time-of-Flight MS, Agilent) , Positive | splash10-004i-9000000000-c38d0fb28793438083a9 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - DI-ESI-Q-Exactive Plus , Positive | splash10-004i-9000000000-18a7ae48c7b0e15cdf18 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-00di-9000000000-6001578fc511ba3fefef | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-00di-9000000000-79b2a0a9d93de6a62358 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-00di-9000000000-9290dbe208c4744f4431 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , negative | splash10-00di-9000000000-605b44ac311a9af4bb7a | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-004i-9000000000-342ab462db0835abb3d2 | 2017-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0059-9000000000-c6b1ebc1dba89b6a6184 | 2015-04-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9000000000-851aa6a0263541a8b249 | 2015-04-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-053r-9000000000-d3b5624412082bb2cf60 | 2015-04-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-9000000000-89b2c043a5afe3ebc6f6 | 2015-04-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-9000000000-b4046e208ee8adb87021 | 2015-04-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05fr-9000000000-36521e440c602bd2ca5a | 2015-04-25 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 22.5 MHz, D2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Ammonia Recycling |  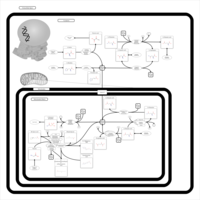 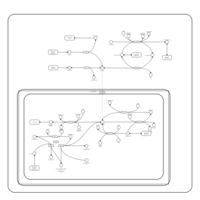 |  | | Bile Acid Biosynthesis | 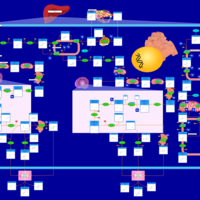 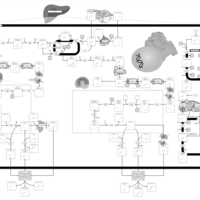  | 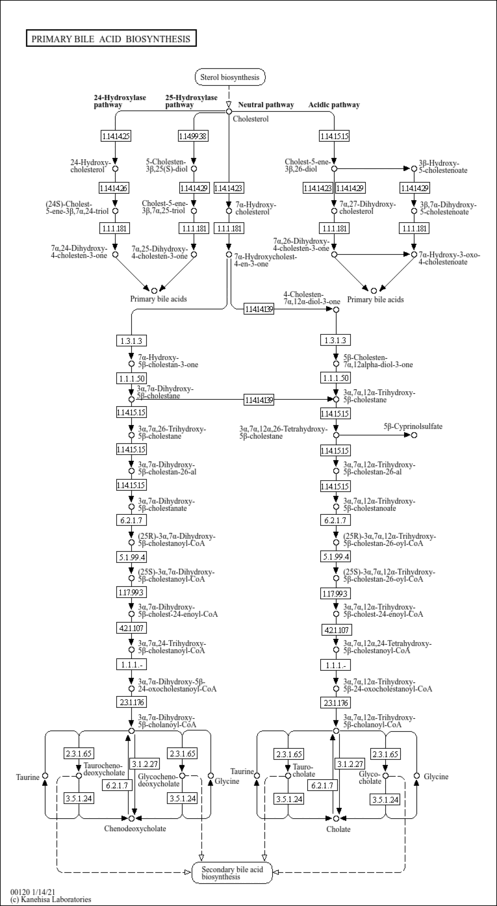 | | Porphyrin Metabolism |   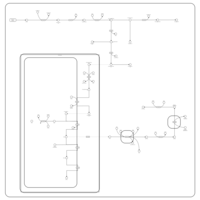 |  | | Glutathione Metabolism | 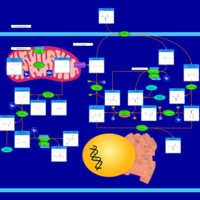 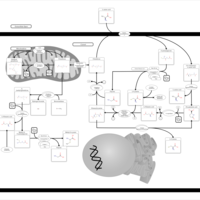 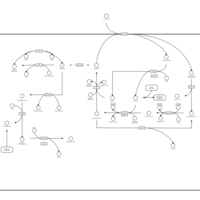 |  | | Glycine and Serine Metabolism | 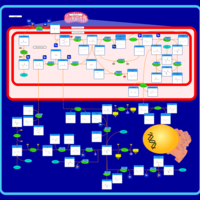 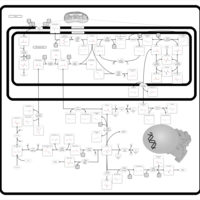 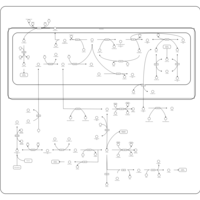 |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| 5-aminolevulinate synthase, erythroid-specific, mitochondrial | ALAS2 | Xp11.21 | P22557 | details | | 5-aminolevulinate synthase, nonspecific, mitochondrial | ALAS1 | 3p21.1 | P13196 | details | | 2-amino-3-ketobutyrate coenzyme A ligase, mitochondrial | GCAT | 22q13.1 | O75600 | details | | Peroxisomal sarcosine oxidase | PIPOX | 17q11.2 | Q9P0Z9 | details | | D-amino-acid oxidase | DAO | 12q24 | P14920 | details | | Trifunctional purine biosynthetic protein adenosine-3 | GART | 21q22.11 | P22102 | details | | Alanine--glyoxylate aminotransferase 2, mitochondrial | AGXT2 | 5p13 | Q9BYV1 | details | | Glycine N-methyltransferase | GNMT | 6p12 | Q14749 | details | | Aminopeptidase N | ANPEP | | P15144 | details | | Glycine amidinotransferase, mitochondrial | GATM | 15q21.1 | P50440 | details | | Bile acid-CoA:amino acid N-acyltransferase | BAAT | 9q22.3 | Q14032 | details | | Glycine--tRNA ligase | GARS | 7p15 | P41250 | details | | Aminomethyltransferase, mitochondrial | AMT | 3p21.2-p21.1 | P48728 | details | | Serine--pyruvate aminotransferase | AGXT | 2q37.3 | P21549 | details | | Glutathione synthetase | GSS | 20q11.2 | P48637 | details | | Serine hydroxymethyltransferase, mitochondrial | SHMT2 | 12q12-q14 | P34897 | details | | Sarcosine dehydrogenase, mitochondrial | SARDH | 9q33-q34 | Q9UL12 | details | | Serine hydroxymethyltransferase, cytosolic | SHMT1 | 17p11.2 | P34896 | details | | Glycine dehydrogenase [decarboxylating], mitochondrial | GLDC | 9p22 | P23378 | details | | Cytosol aminopeptidase | LAP3 | | P28838 | details | | Glycine cleavage system H protein, mitochondrial | GCSH | | P23434 | details | | Vesicular inhibitory amino acid transporter | SLC32A1 | 20q11.23 | Q9H598 | details | | Glycine N-acyltransferase | GLYAT | 11q12.1 | Q6IB77 | details | | Glycine N-acyltransferase-like protein 1 | GLYATL1 | 11q12.1 | Q969I3 | details | | Glycine N-acyltransferase-like protein 2 | GLYATL2 | 11q12.1 | Q8WU03 | details | | Kynurenine--oxoglutarate transaminase 3 | CCBL2 | 1p22.2 | Q6YP21 | details |
|
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 0.137 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 0.0932 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 0.0520 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.177 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 0.104 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 0.0849 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000123 |
|---|
| DrugBank ID | DB00145 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB000484 |
|---|
| KNApSAcK ID | C00001361 |
|---|
| Chemspider ID | 730 |
|---|
| KEGG Compound ID | C00037 |
|---|
| BioCyc ID | GLY |
|---|
| BiGG ID | 33610 |
|---|
| Wikipedia Link | Glycine |
|---|
| METLIN ID | 20 |
|---|
| PubChem Compound | 750 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15428 |
|---|
| References |
|---|
| General References | - Razak MA, Begum PS, Viswanath B, Rajagopal S: Multifarious Beneficial Effect of Nonessential Amino Acid, Glycine: A Review. Oxid Med Cell Longev. 2017;2017:1716701. doi: 10.1155/2017/1716701. Epub 2017 Mar 1. [PubMed:28337245 ]
- McCarty MF, DiNicolantonio JJ: The cardiometabolic benefits of glycine: Is glycine an 'antidote' to dietary fructose? Open Heart. 2014 May 28;1(1):e000103. doi: 10.1136/openhrt-2014-000103. eCollection 2014. [PubMed:25332814 ]
- Perez-Torres I, Zuniga-Munoz AM, Guarner-Lans V: Beneficial Effects of the Amino Acid Glycine. Mini Rev Med Chem. 2017;17(1):15-32. doi: 10.2174/1389557516666160609081602. [PubMed:27292783 ]
|
|---|




