| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-19 00:46:25 UTC |
|---|
| Updated at | 2020-12-07 19:07:34 UTC |
|---|
| CannabisDB ID | CDB000608 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Formic acid |
|---|
| Description | Formic acid, also known as methanoic acid, is the simplest carboxylic acid, and has the chemical formula HCOOH. It is an important intermediate in chemical synthesis and occurs naturally, most notably in some ants. Esters, salts, and the anion derived from formic acid are called formates. Industrially, formic acid is produced through the oxidation of methanol. Formate is an intermediate in normal metabolism. It takes part in the metabolism of one-carbon compounds and its carbon may appear in methyl groups undergoing transmethylation. It is eventually oxidized to carbon dioxide. Formate is typically produced as a by-product in the production of acetate. It is responsible for both metabolic acidosis and disrupting mitochondrial electron transport and energy production by inhibiting cytochrome oxidase activity, the terminal electron acceptor of the electron transport chain. Cell death from cytochrome oxidase inhibition by formate is believed to result partly from depletion of ATP, reducing energy concentrations so that essential cell functions cannot be maintained. Furthermore, inhibition of cytochrome oxidase by formate may also cause cell death by increased production of cytotoxic reactive oxygen species (ROS) secondary to the blockade of the electron transport chain. In nature, formic acid is found in the stings and bites of many insects of the order Hymenoptera, including bees and ants. The principal use of formic acid is as a preservative and antibacterial agent in livestock feed. When sprayed on fresh hay or other silage, it arrests certain decay processes and causes the feed to retain its nutritive value longer. Urinary formate is produced by Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumonia, Enterobacter, Acinetobacter, Proteus mirabilis, Citrobacter frundii, Enterococcus faecalis, Streptococcus group B, Staphylococcus saprophyticus (PMID: 22292465 ). Formic acid has also been detected as a volatile component in cannabis plant samples (PMID: 26657499 ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Acide formique | ChEBI | | Ameisensaeure | ChEBI | | Aminic acid | ChEBI | | Bilorin | ChEBI | | Formylic acid | ChEBI | | H-COOH | ChEBI | | HCO2H | ChEBI | | HCOOH | ChEBI | | Hydrogen carboxylic acid | ChEBI | | Methanoic acid | ChEBI | | Methoic acid | ChEBI | | Aminate | Generator | | Formylate | Generator | | Hydrogen carboxylate | Generator | | Methanoate | Generator | | Methoate | Generator | | Formate | Generator | | Add-F | HMDB | | Ameisensaure | HMDB | | Collo-bueglatt | HMDB | | Collo-didax | HMDB | | Formira | HMDB | | Formisoton | HMDB | | Methanoic acid monomer | HMDB | | Myrmicyl | HMDB | | Sodium formate | HMDB | | Sybest | HMDB | | Wonderbond hardener m 600l | HMDB | | Calcium formate | HMDB | | Cobalt(II) formate dihydrate | HMDB | | Formic acid, aluminum salt | HMDB | | Formic acid, copper salt | HMDB | | Formic acid, cromium (+3) salt | HMDB | | Lithium formate | HMDB | | Ammonium formate | HMDB | | Formic acid, ammonium (4:1) salt | HMDB | | Formic acid, ammonium salt | HMDB | | Formic acid, calcium salt | HMDB | | Formic acid, copper (+2) salt | HMDB | | Formic acid, lead (+2) salt | HMDB | | Formic acid, lead salt | HMDB | | Formic acid, nickel salt | HMDB | | Formic acid, potassium salt | HMDB | | Formic acid, strontium salt | HMDB | | Mafusol | HMDB | | Ammonium tetraformate | HMDB | | Formic acid, 14C-labeled | HMDB | | Formic acid, cobalt (+2) salt | HMDB | | Formic acid, copper, ammonium salt | HMDB | | Formic acid, sodium salt | HMDB | | Formic acid, sodium salt, 14C-labeled | HMDB | | Formic acid, ammonium (2:1) salt | HMDB | | Formic acid, cadmium salt | HMDB | | Formic acid, cesium salt | HMDB | | Formic acid, copper, nickel salt | HMDB | | Formic acid, cromium (+3), sodium (4:1:1) salt | HMDB | | Formic acid, lithium salt | HMDB | | Formic acid, magnesium salt | HMDB | | Formic acid, nickel (+2) salt | HMDB | | Formic acid, rubidium salt | HMDB | | Formic acid, sodium salt, 13C-labeled | HMDB | | Formic acid, thallium (+1) salt | HMDB | | Formic acid, zinc salt | HMDB | | Nickel formate dihydrate | HMDB | | Aluminum formate | HMDB | | Potassium formate | HMDB | | Strontium formate | HMDB | | Lead formate | HMDB | | Nickel formate | HMDB | | Chromic formate | HMDB | | Cobaltous formate | HMDB | | Cupric formate | HMDB | | Magnesium formate | HMDB | | Zinc formate | HMDB |
|
|---|
| Chemical Formula | CH2O2 |
|---|
| Average Molecular Weight | 46.03 |
|---|
| Monoisotopic Molecular Weight | 46.0055 |
|---|
| IUPAC Name | formic acid |
|---|
| Traditional Name | formic acid |
|---|
| CAS Registry Number | 64-18-6 |
|---|
| SMILES | OC=O |
|---|
| InChI Identifier | InChI=1S/CH2O2/c2-1-3/h1H,(H,2,3) |
|---|
| InChI Key | BDAGIHXWWSANSR-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as carboxylic acids. Carboxylic acids are compounds containing a carboxylic acid group with the formula -C(=O)OH. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Carboxylic acids |
|---|
| Direct Parent | Carboxylic acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Monocarboxylic acid or derivatives
- Carboxylic acid
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Environmental role: Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 8.4 °C | Not Available | | Boiling Point | 100.8 °C | Wikipedia | | Water Solubility | 1000 mg/mL | Not Available | | logP | -0.54 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-004j-9000000000-2e63b0c1e2e417b0d747 | 2015-03-01 | View Spectrum | | Predicted GC-MS | Formic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0002-9000000000-5d27bb312e37a2c8994f | Spectrum | | Predicted GC-MS | Formic acid, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0fmi-9200000000-2a89ba98485194acd75a | Spectrum | | Predicted GC-MS | Formic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Formic acid, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - QqQ 4V, positive | splash10-0002-9000000000-a8fbddf8ca4197b30013 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 5V, positive | splash10-0002-9000000000-98310116f8a2d6a969ce | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 6V, positive | splash10-0002-9000000000-ec8753fd9790a3cf0be8 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 7V, positive | splash10-0002-9000000000-121d1a025b72a70e412a | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 8V, positive | splash10-0002-9000000000-5f1955dee7ab988e86dd | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 9V, positive | splash10-0002-9000000000-f8e14272296ee06d19f4 | 2020-07-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9000000000-092f816e62c8d2f5d56e | 2015-04-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9000000000-092f816e62c8d2f5d56e | 2015-04-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9000000000-092f816e62c8d2f5d56e | 2015-04-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9000000000-eb2207f7400e9144fff7 | 2015-04-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-eb2207f7400e9144fff7 | 2015-04-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-eb2207f7400e9144fff7 | 2015-04-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9000000000-d948d5d95ae14e701f57 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-d948d5d95ae14e701f57 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-d948d5d95ae14e701f57 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9000000000-92190863fc6ae28a8789 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9000000000-92190863fc6ae28a8789 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9000000000-348f481062f48991a0aa | 2021-09-22 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 15.09 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Pterine Biosynthesis |    | 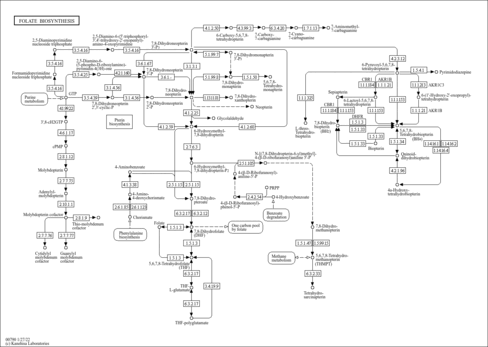 | | Folate Metabolism |    | 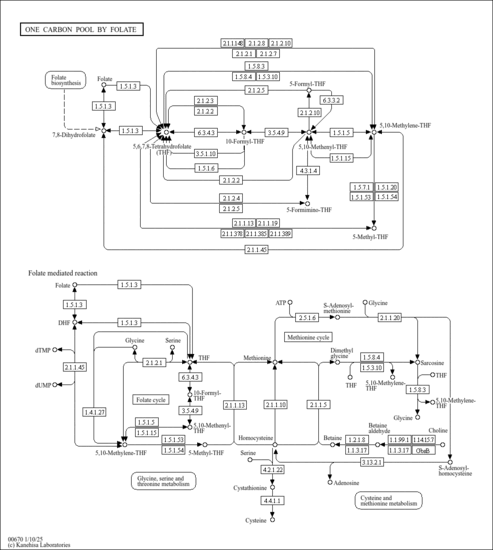 | | Methotrexate Action Pathway | 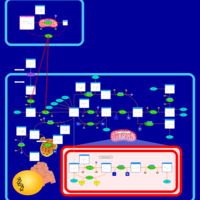 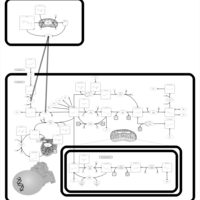 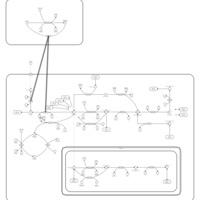 | Not Available | | Folate malabsorption, hereditary | 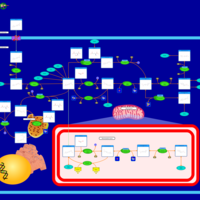 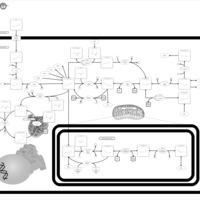 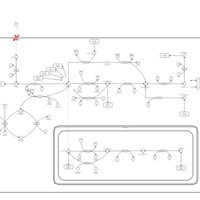 | Not Available | | Steroid Biosynthesis | 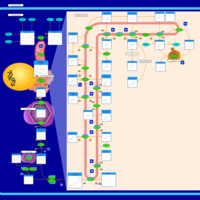 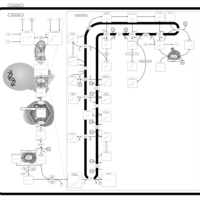  |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Lanosterol 14-alpha demethylase | CYP51A1 | 7q21.2 | Q16850 | details | | S-formylglutathione hydrolase | ESD | 13q14.1-q14.2 | P10768 | details | | Aspartoacylase | ASPA | 17p13.3 | P45381 | details | | Aspartoacylase-2 | ACY3 | | Q96HD9 | details | | C-1-tetrahydrofolate synthase, cytoplasmic | MTHFD1 | 14q24 | P11586 | details | | GTP cyclohydrolase 1 | GCH1 | 14q22.1-q22.2 | P30793 | details | | Peptide deformylase, mitochondrial | PDF | 16q22.1 | Q9HBH1 | details | | Monofunctional C1-tetrahydrofolate synthase, mitochondrial | MTHFD1L | 6q25.1 | Q6UB35 | details | | Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase | MTHFD2 | 2p13.1 | Q7Z650 | details | | Kynurenine formamidase | AFMID | 17q25.3 | Q63HM1 | details | | Cytochrome P450, family 1, subfamily A, polypeptide 1 | CYP1A1 | 15q24.1 | A0N0X8 | details | | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase | ADI1 | 2p25.3 | Q9BV57 | details |
|
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000142 |
|---|
| DrugBank ID | DB01942 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB012804 |
|---|
| KNApSAcK ID | C00001182 |
|---|
| Chemspider ID | Not Available |
|---|
| KEGG Compound ID | C00058 |
|---|
| BioCyc ID | FORMATE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Formic_acid |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 284 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 30751 |
|---|
| References |
|---|
| General References | - Gupta A, Dwivedi M, Mahdi AA, Khetrapal CL, Bhandari M: Broad identification of bacterial type in urinary tract infection using (1)h NMR spectroscopy. J Proteome Res. 2012 Mar 2;11(3):1844-54. doi: 10.1021/pr2010692. Epub 2012 Jan 31. [PubMed:22292465 ]
- Rice S, Koziel JA: Characterizing the Smell of Marijuana by Odor Impact of Volatile Compounds: An Application of Simultaneous Chemical and Sensory Analysis. PLoS One. 2015 Dec 10;10(12):e0144160. doi: 10.1371/journal.pone.0144160. eCollection 2015. [PubMed:26657499 ]
|
|---|


