| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-27 16:26:41 UTC |
|---|
| Updated at | 2021-01-04 20:37:45 UTC |
|---|
| CannabisDB ID | CDB005499 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Nicotine |
|---|
| Description | Nicotine, also known as (S)-nicotine or habitrol, belongs to the class of organic compounds known as pyrrolidinylpyridines. Pyrrolidinylpyridines are compounds containing a pyrrolidinylpyridine ring system, which consists of a pyrrolidine ring linked to a pyridine ring. Nicotine is a hygroscopic, oily liquid that is miscible with water in its base form. As a nitrogenous base, nicotine forms salts with acids that are usually solid and water soluble. Nicotine is an alkaloid found in the nightshade family of plants (Solanaceae), predominantly in tobacco and in lower quantities in tomato, potato, eggplant (aubergine), and green pepper. Nicotine alkaloids are also found in the leaves of the coca plant. Nicotine constitutes 0.3 to 5% of the tobacco plant by dry weight, with biosynthesis taking place in the root and accumulation in the leaves. Nicotine is highly addictive and it is the main reason why tobacco smoking (or chewing tobacco) leads to life-long addiction or dependency in many individuals. Nicotine causes the release dopamine in the same regions of the brain as other addictive drugs. It causes mood-altering changes that make the person temporarily feel good. Inhaled tobacco smoke delivers nicotine to the brain within 20 seconds, which makes tobacco smoke (and smoking) very addictive—comparable to other addictive drugs such as opioids, alcohol and cocaine. The amount of nicotine inhaled with tobacco smoke is a fraction of the amount contained in the tobacco leaves (most of the substance is destroyed by the heat). The amount of nicotine absorbed by the body from smoking depends on many factors, including the type of tobacco, whether the smoke is inhaled, and whether a filter is used. For chewing tobacco, often called dip, snuff, or sinus, which is held in the mouth between the lip and gum, the amount released into the body tends to be much greater than smoked tobacco. Nicotine is a stimulant that acts as an agonist at nicotinic acetylcholine receptors. These are ionotropic receptors composed of five homomeric or heteromeric subunits. In the brain, nicotine binds to nicotinic acetylcholine receptors on dopaminergic neurons in the cortico-limbic pathways. This causes the channel to open and allow conductance of multiple cations including sodium, calcium, and potassium. This leads to depolarization, which activates voltage-gated calcium channels and allows more calcium to enter the axon terminal. Calcium stimulates vesicle trafficking towards the plasma membrane and the release of dopamine into the synapse. Dopamine binding to its receptors is responsible for the euphoric and addictive properties of nicotine. Nicotine also binds to nicotinic acetylcholine receptors on the chromaffin cells in the adrenal medulla. Binding opens the ion channel allowing an influx of sodium which causes depolarization of the cell and activates voltage-gated calcium channels. Calcium triggers the release of epinephrine from intracellular vesicles into the bloodstream, which causes vasoconstriction, increased blood pressure, increased heart rate, and increased blood sugar. It has been noted that the majority of people diagnosed with schizophrenia smoke tobacco. Estimates for the number of schizophrenics that smoke range from 75% to 90%. It was recently argued that the increased level of smoking in schizophrenia may be due to a desire to self-medicate with nicotine. Nicotine is a potent neurotoxin with particular specificity to insects; therefore nicotine was widely used as an insecticide in the past and nicotine derivatives such as imidacloprid continue to be widely used as an insecticide. Nicotine is not a carcinogent and nicotine on its own, does not promote the development of cancer in healthy tissue and has no mutagenic properties. However, nicotine and the increased acetylcholinic activity it causes have been shown to impede apoptosis, which is one of the methods by which the body destroys unwanted cells (programmed cell death). Since apoptosis helps to remove mutated or damaged cells that may eventually become cancerous, the inhibitory actions of nicotine create a more favourable environment for cancer to develop. Nicotine is also a constituent of cannabis smoke. It is formed during the combustion of cannabis ( Ref:DOI ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (-)-Nicotine | ChEBI | | (-)-3-(1-Methyl-2-pyrrolidyl)pyridine | ChEBI | | (-)-3-(N-Methylpyrrolidino)pyridine | ChEBI | | (R)-3-(1-Methyl-2-pyrrolidinyl)pyridine | ChEBI | | (S)-(-)-Nicotine | ChEBI | | (S)-3-(1-Methylpyrrolidin-2-yl)pyridine | ChEBI | | (S)-3-(N-Methylpyrrolidin-2-yl)pyridine | ChEBI | | 1-Methyl-2-(3-pyridyl)pyrrolidine | ChEBI | | 3-(1-Methyl-2-pyrollidinyl)pyridine | ChEBI | | 3-(1-Methylpyrrolidin-2-yl)pyridine | ChEBI | | 3-(2-(N-Methylpyrrolidinyl))pyridine | ChEBI | | 3-(N-Methylpyrollidino)pyridine | ChEBI | | L(-)-Nicotine | ChEBI | | L-3-(1-Methyl-2-pyrrolidyl)pyridine | ChEBI | | L-Nicotine | ChEBI | | (S)-Nicotine | Kegg | | Habitrol | Kegg | | (+)-Nicotine | HMDB | | (R,S)-Nicotine | HMDB | | 1-Methyl-2-(3-pyridal)-pyrrolidene | HMDB | | 1-Methyl-2-(3-pyridal)-pyrrolidine | HMDB | | 1-Methyl-2-(3-pyridiyl)pyrrolidine | HMDB | | 2'-beta-H-Nicotine | HMDB | | 3-(1-Methyl-2-pyrrolidinyl)-pyridine | HMDB | | 3-(1-Methyl-2-pyrrolidinyl)pyridine | HMDB | | 3-(N-Methylpyrrolidino)pyridine | HMDB | | 3-N-Methylpyrrolidine | HMDB | | a -N-Methylpyrrolidine | HMDB | | a-N-Methylpyrrolidine | HMDB | | alpha-N-Methylpyrrolidine | HMDB | | beta-Pyridyl-alpha-N-methylpyrrolidine | HMDB | | D-Nicotine | HMDB | | delta-Nicotine | HMDB | | Destruxol | HMDB | | DL-Tetrahydronicotyrine | HMDB | | Fumeto bac | HMDB | | Methyl-2-pyrrolidinyl)pyridine | HMDB | | Nicoderm | HMDB | | Nicotine polacrilex | HMDB | | R)-(+)-Nicotine | HMDB | | Tartrate, nicotine | HMDB | | Nicotine bitartrate | HMDB | | Nicotine tartrate | HMDB | | Bitartrate, nicotine | HMDB | | Nicotine | ChEBI |
|
|---|
| Chemical Formula | C10H14N2 |
|---|
| Average Molecular Weight | 162.23 |
|---|
| Monoisotopic Molecular Weight | 162.1157 |
|---|
| IUPAC Name | 3-[(2S)-1-methylpyrrolidin-2-yl]pyridine |
|---|
| Traditional Name | nicoderm CQ |
|---|
| CAS Registry Number | 54-11-5 |
|---|
| SMILES | CN1CCC[C@H]1C1=CN=CC=C1 |
|---|
| InChI Identifier | InChI=1S/C10H14N2/c1-12-7-3-5-10(12)9-4-2-6-11-8-9/h2,4,6,8,10H,3,5,7H2,1H3/t10-/m0/s1 |
|---|
| InChI Key | SNICXCGAKADSCV-JTQLQIEISA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as pyrrolidinylpyridines. Pyrrolidinylpyridines are compounds containing a pyrrolidinylpyridine ring system, which consists of a pyrrolidine ring linked to a pyridine ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pyridines and derivatives |
|---|
| Sub Class | Pyrrolidinylpyridines |
|---|
| Direct Parent | Pyrrolidinylpyridines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrrolidinylpyridine
- Alkaloid or derivatives
- Aralkylamine
- N-alkylpyrrolidine
- Heteroaromatic compound
- Pyrrolidine
- Tertiary aliphatic amine
- Tertiary amine
- Azacycle
- Organic nitrogen compound
- Organopnictogen compound
- Hydrocarbon derivative
- Organonitrogen compound
- Amine
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Environmental role: Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Liquid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | -79 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 1000 mg/mL | Not Available | | logP | 1.17 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-001i-9400000000-47a036aa305825218fa2 | 2014-09-20 | View Spectrum | | GC-MS | Nicotine, non-derivatized, GC-MS Spectrum | splash10-001i-9500000000-f2d4835c504301f9410e | Spectrum | | GC-MS | Nicotine, non-derivatized, GC-MS Spectrum | splash10-01q9-7900000000-1e2c38b5e4e7aae10d37 | Spectrum | | GC-MS | Nicotine, non-derivatized, GC-MS Spectrum | splash10-001i-9800000000-fe889308f7088d47c31d | Spectrum | | GC-MS | Nicotine, non-derivatized, GC-MS Spectrum | splash10-001i-9500000000-f2d4835c504301f9410e | Spectrum | | Predicted GC-MS | Nicotine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-001i-4900000000-6d65165a4417a6129eeb | Spectrum | | Predicted GC-MS | Nicotine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-00di-0900000000-3e7377f36ca2547f4885 | 2012-07-25 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001i-2900000000-a505fff3a4028a6f0d52 | 2012-07-25 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00lr-7900000000-b91a12a16d7688e8679d | 2012-07-25 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI M-80) , Positive | splash10-01q9-7900000000-10d401d7ffa1c2cdbd9f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-03di-0900000000-440798524836a27b78b4 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-01q9-0900000000-926940dd7f4851dbd73b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-00lr-0900000000-6bccc06d40ab273cf972 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-0159-2900000000-f7512c405bb662e040a6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-014i-7900000000-f1781566be5aee88f6ef | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-001i-0900000000-09e9b29571abf3a52e44 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-03di-0900000000-16cdb5f23105be4a892b | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-03di-0900000000-720fe01876c739a30be0 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-001i-0900000000-85a813588b57acf531fe | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-001i-0900000000-93a3dae1577f4f2dda53 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-001i-1900000000-cb6f8cec40dd9ed35d00 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-00lr-1900000000-1443b1cf22d2de281898 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-03di-0900000000-440798524836a27b78b4 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-01q9-0900000000-926940dd7f4851dbd73b | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-00lr-0900000000-6bccc06d40ab273cf972 | 2017-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0900000000-bf73c660c9fef052a3e2 | 2017-07-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-1900000000-317ad58e8bc4c3092420 | 2017-07-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-053u-9400000000-ea4895202e26a55d8a9b | 2017-07-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0900000000-ed813017157d07798ea2 | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-1900000000-02ba66ba693d048f19ab | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9400000000-442e757e1da476d883af | 2017-07-26 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 15.09 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Nicotine Action Pathway | 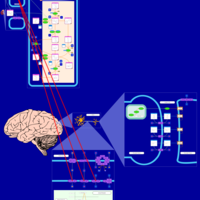 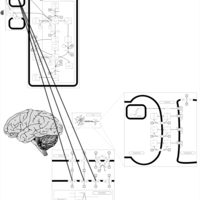  | Not Available | | Nicotine Metabolism Pathway | 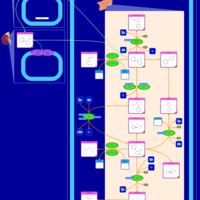 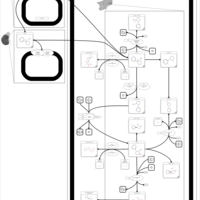 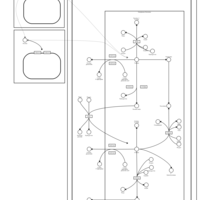 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | |
| Neuronal acetylcholine receptor subunit alpha-10 | CHRNA10 | 11p15.5 | Q9GZZ6 | details | | Neuronal acetylcholine receptor subunit alpha-9 | CHRNA9 | 4p14 | Q9UGM1 | details | | Neuronal acetylcholine receptor subunit alpha-2 | CHRNA2 | 8p21 | Q15822 | details |
|
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0001934 |
|---|
| DrugBank ID | DB00184 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB003968 |
|---|
| KNApSAcK ID | C00002057 |
|---|
| Chemspider ID | 80863 |
|---|
| KEGG Compound ID | C00745 |
|---|
| BioCyc ID | NICOTINE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Nicotine |
|---|
| METLIN ID | 1526 |
|---|
| PubChem Compound | 89594 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17688 |
|---|
| References |
|---|
| General References | Not Available |
|---|