| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:36:00 UTC |
|---|
| Updated at | 2020-11-18 16:38:39 UTC |
|---|
| CannabisDB ID | CDB004780 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | 2-Ketobutyric acid |
|---|
| Description | 2-Ketobutyric acid, also known as alpha-ketobutyrate or 2-oxobutyrate, belongs to the class of organic compounds known as short-chain keto acids and derivatives. These are keto acids with an alkyl chain the contains less than 6 carbon atoms. 2-Ketobutyric acid is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. 2-Ketobutyric acid exists in all living species, ranging from bacteria to humans. Within humans, 2-ketobutyric acid participates in a number of enzymatic reactions. In particular, 2-ketobutyric acid can be biosynthesized from L-threonine; which is mediated by the enzyme L-serine dehydratase/l-threonine deaminase. In addition, 2-ketobutyric acid and coenzyme A can be converted into propionyl-CoA; which is mediated by the enzyme branched-chain alpha-keto dehydrogenase complex. In humans, 2-ketobutyric acid is involved in threonine and 2-oxobutanoate degradation. Outside of the human body, 2-Ketobutyric acid has been detected, but not quantified in, several different foods, such as black crowberries, lima beans, lettuces, oxheart cabbages, and grass pea. This could make 2-ketobutyric acid a potential biomarker for the consumption of these foods. A 2-oxo monocarboxylic acid that is the 2-oxo derivative of butanoic acid. 2-Ketobutyric acid is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Ketobutanoic acid | ChEBI | | 2-Oxobutyric acid | ChEBI | | 3-Methyl pyruvic acid | ChEBI | | alpha-Ketobutyrate | ChEBI | | alpha-Ketobutyric acid | ChEBI | | alpha-oxo-N-Butyric acid | ChEBI | | 2-Oxobutyrate | Kegg | | 2-Oxobutanoic acid | Kegg | | 2-Ketobutanoate | Generator | | 3-Methyl pyruvate | Generator | | a-Ketobutyrate | Generator | | a-Ketobutyric acid | Generator | | Α-ketobutyrate | Generator | | Α-ketobutyric acid | Generator | | a-oxo-N-Butyrate | Generator | | a-oxo-N-Butyric acid | Generator | | alpha-oxo-N-Butyrate | Generator | | Α-oxo-N-butyrate | Generator | | Α-oxo-N-butyric acid | Generator | | 2-Oxobutanoate | Generator | | 2-Ketobutyrate | Generator | | 2-oxo-Butanoate | HMDB | | 2-oxo-Butanoic acid | HMDB | | 2-oxo-Butyrate | HMDB | | 2-oxo-Butyric acid | HMDB | | 2-oxo-N-Butyrate | HMDB | | 2-oxo-N-Butyric acid | HMDB | | 3-Methylpyruvate | HMDB | | 3-Methylpyruvic acid | HMDB | | a-Keto-N-butyrate | HMDB | | a-Keto-N-butyric acid | HMDB | | a-Oxobutyrate | HMDB | | a-Oxobutyric acid | HMDB | | alpha-Keto-N-butyrate | HMDB | | alpha-Keto-N-butyric acid | HMDB | | alpha-Ketobutric acid | HMDB | | alpha-Oxobutyrate | HMDB | | alpha-Oxobutyric acid | HMDB | | Methyl-pyruvate | HMDB | | Methyl-pyruvic acid | HMDB | | Propionyl-formate | HMDB | | Propionyl-formic acid | HMDB | | alpha-Ketobutyric acid, sodium salt | HMDB | | 2-Ketobutyric acid | ChEBI |
|
|---|
| Chemical Formula | C4H6O3 |
|---|
| Average Molecular Weight | 102.09 |
|---|
| Monoisotopic Molecular Weight | 102.0317 |
|---|
| IUPAC Name | 2-oxobutanoic acid |
|---|
| Traditional Name | 2-oxobutanoic acid |
|---|
| CAS Registry Number | 600-18-0 |
|---|
| SMILES | CCC(=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C4H6O3/c1-2-3(5)4(6)7/h2H2,1H3,(H,6,7) |
|---|
| InChI Key | TYEYBOSBBBHJIV-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as short-chain keto acids and derivatives. These are keto acids with an alkyl chain the contains less than 6 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Keto acids and derivatives |
|---|
| Sub Class | Short-chain keto acids and derivatives |
|---|
| Direct Parent | Short-chain keto acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Short-chain keto acid
- Alpha-keto acid
- Alpha-hydroxy ketone
- Ketone
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 33 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 119 mg/mL | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | 2-Ketobutyric acid, non-derivatized, GC-MS Spectrum | splash10-0gbj-1900000000-8daa299f5bc38a7f9b3e | Spectrum | | GC-MS | 2-Ketobutyric acid, non-derivatized, GC-MS Spectrum | splash10-014s-1910000000-d01bcd7b41fdcc634eea | Spectrum | | GC-MS | 2-Ketobutyric acid, 1 MEOX; 1 TMS, GC-MS Spectrum | splash10-000i-9200000000-3b3816ffee5771215904 | Spectrum | | GC-MS | 2-Ketobutyric acid, 1 MEOX; 1 TMS, GC-MS Spectrum | splash10-000i-6910000000-5b264d5f7e53f3e80b12 | Spectrum | | GC-MS | 2-Ketobutyric acid, 2 TMS, GC-MS Spectrum | splash10-001i-6950000000-db7f6a0f03a8cce59bae | Spectrum | | GC-MS | 2-Ketobutyric acid, non-derivatized, GC-MS Spectrum | splash10-0ftb-0920000000-a6c93d8761ef15f7f7d4 | Spectrum | | GC-MS | 2-Ketobutyric acid, non-derivatized, GC-MS Spectrum | splash10-014i-0920000000-406fe70e72dbd9c3f274 | Spectrum | | GC-MS | 2-Ketobutyric acid, non-derivatized, GC-MS Spectrum | splash10-0gbj-1900000000-8daa299f5bc38a7f9b3e | Spectrum | | GC-MS | 2-Ketobutyric acid, non-derivatized, GC-MS Spectrum | splash10-014s-1910000000-d01bcd7b41fdcc634eea | Spectrum | | GC-MS | 2-Ketobutyric acid, non-derivatized, GC-MS Spectrum | splash10-000i-9200000000-3b3816ffee5771215904 | Spectrum | | GC-MS | 2-Ketobutyric acid, non-derivatized, GC-MS Spectrum | splash10-000i-6910000000-5b264d5f7e53f3e80b12 | Spectrum | | GC-MS | 2-Ketobutyric acid, non-derivatized, GC-MS Spectrum | splash10-001i-6950000000-db7f6a0f03a8cce59bae | Spectrum | | GC-MS | 2-Ketobutyric acid, non-derivatized, GC-MS Spectrum | splash10-000i-8900000000-97bab6630de4df095a7f | Spectrum | | GC-MS | 2-Ketobutyric acid, non-derivatized, GC-MS Spectrum | splash10-000i-9200000000-0ebb2354fe3f935fe177 | Spectrum | | Predicted GC-MS | 2-Ketobutyric acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0a6r-9000000000-32269ef3ef778c482f99 | Spectrum | | Predicted GC-MS | 2-Ketobutyric acid, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0adi-9200000000-f92941e9f326d8ccf3e4 | Spectrum | | Predicted GC-MS | 2-Ketobutyric acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | 2-Ketobutyric acid, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | 2-Ketobutyric acid, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | 2-Ketobutyric acid, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-0pb9-9600000000-53ebb7164f74d7d92a4d | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-0zfr-5900000000-1db864754705beb79b55 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-0udi-9800000000-5ba5b9ede5b1bc389b87 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0udi-0900000000-bfdcad5bc3ed586bdede | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0udi-0900000000-bfdcad5bc3ed586bdede | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 4V, positive | splash10-0pb9-9600000000-b10635fbec5572d4d084 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 8V, positive | splash10-0a4i-9100000000-4c5e7d24dca6677e1570 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 12V, positive | splash10-0a4i-9000000000-ded44cfcbe7960954fdf | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 16V, positive | splash10-0a4i-9000000000-1b350be7438b08f1946a | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 20V, positive | splash10-056r-9000000000-172a175b8b9ebfd5529f | 2020-07-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0k9i-9400000000-0061f4635858d851f127 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4r-9100000000-b943445194fb5f8e3a0f | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-bc5920b9bd140be6b06d | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-5900000000-f7dfb4554f0b9e1499c9 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a59-9200000000-e54106ad5bf45a9f072c | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9000000000-7bbba57578cf720974e3 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-9000000000-ca1ed9c5aadcbdabc2a3 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-9000000000-aced8e004436190f07b7 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-a3d7ddf227bd6aa91a54 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0pb9-9700000000-503296025148ec314383 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-9000000000-933b567955b5de5b1744 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-7d9db52021fefc87a855 | 2021-09-25 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Methionine Metabolism | 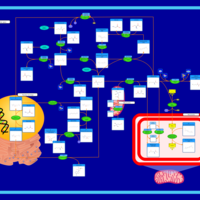 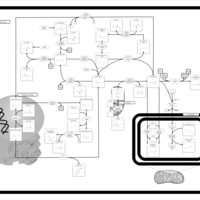 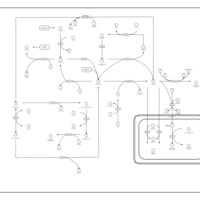 |  | | Glycine and Serine Metabolism | 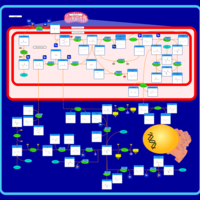 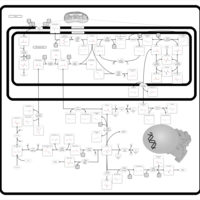 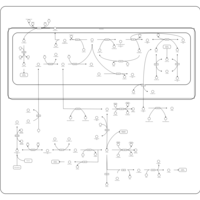 |  | | Threonine and 2-Oxobutanoate Degradation | 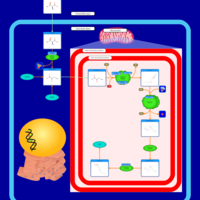 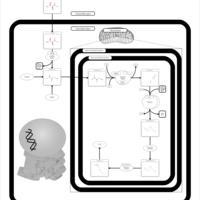 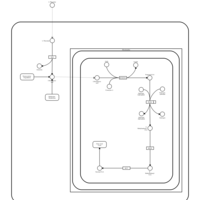 | Not Available | | Homocysteine Degradation |   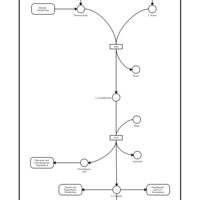 | Not Available | | Selenoamino Acid Metabolism |  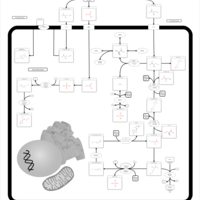  |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | PDHB | 3p21.1-p14.2 | P11177 | details | | Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial | PDHA1 | Xp22.1 | P08559 | details | | Pyruvate dehydrogenase E1 component subunit alpha, testis-specific form, mitochondrial | PDHA2 | 4q22-q23 | P29803 | details | | 2-oxoisovalerate dehydrogenase subunit beta, mitochondrial | BCKDHB | 6q14.1 | P21953 | details | | 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial | BCKDHA | 19q13.1-q13.2 | P12694 | details | | 2-oxoglutarate dehydrogenase, mitochondrial | OGDH | 7p14-p13 | Q02218 | details | | Cystathionine gamma-lyase | CTH | 1p31.1 | P32929 | details | | L-serine dehydratase/L-threonine deaminase | SDS | 12q24.13 | P20132 | details | | L-lactate dehydrogenase A-like 6A | LDHAL6A | 11p15.1 | Q6ZMR3 | details | | Urocanate hydratase | UROC1 | 3q21.3 | Q96N76 | details | | [3-methyl-2-oxobutanoate dehydrogenase [lipoamide]] kinase, mitochondrial | BCKDK | 16p11.2 | O14874 | details | | L-lactate dehydrogenase B chain | LDHB | 12p12.2-p12.1 | P07195 | details | | L-lactate dehydrogenase C chain | LDHC | 11p15.1 | P07864 | details | | L-lactate dehydrogenase A chain | LDHA | 11p15.4 | P00338 | details | | L-lactate dehydrogenase A-like 6B | LDHAL6B | 15q22.2 | Q9BYZ2 | details | | Trimethyllysine dioxygenase, mitochondrial | TMLHE | Xq28 | Q9NVH6 | details | | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2 | ALKBH2 | 12q24.11 | Q6NS38 | details | | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 3 | ALKBH3 | 11p11.2 | Q96Q83 | details | | Serine dehydratase-like | SDSL | 12q24.13 | Q96GA7 | details | | 2-oxoglutarate dehydrogenase-like, mitochondrial | OGDHL | | Q9ULD0 | details |
|
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
| 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial | BCKDHA | 19q13.1-q13.2 | P12694 | details | | Trimethyllysine dioxygenase, mitochondrial | TMLHE | Xq28 | Q9NVH6 | details |
|
|---|
| Receptors | |
|---|
| Transcriptional Factors | |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000005 |
|---|
| DrugBank ID | DB04553 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB030356 |
|---|
| KNApSAcK ID | C00019675 |
|---|
| Chemspider ID | 57 |
|---|
| KEGG Compound ID | C00109 |
|---|
| BioCyc ID | 2-OXOBUTANOATE |
|---|
| BiGG ID | 33889 |
|---|
| Wikipedia Link | Alpha-Ketobutyric_acid |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 58 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 30831 |
|---|
| References |
|---|
| General References | Not Available |
|---|


