| Identification |
|---|
| HMDB Protein ID
| CDBP00293 |
| Secondary Accession Numbers
| Not Available |
| Name
| 2-oxoisovalerate dehydrogenase subunit beta, mitochondrial |
| Description
| Not Available |
| Synonyms
|
- BCKDE1B
- BCKDH E1-beta
- Branched-chain alpha-keto acid dehydrogenase E1 component beta chain
|
| Gene Name
| BCKDHB |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in catalytic activity |
| Specific Function
| The branched-chain alpha-keto dehydrogenase complex catalyzes the overall conversion of alpha-keto acids to acyl-CoA and CO(2). It contains multiple copies of three enzymatic components: branched-chain alpha-keto acid decarboxylase (E1), lipoamide acyltransferase (E2) and lipoamide dehydrogenase (E3).
|
| GO Classification
|
| Biological Process |
| response to glucocorticoid stimulus |
| response to cAMP |
| branched-chain amino acid catabolic process |
| cellular nitrogen compound metabolic process |
| response to nutrient |
| Cellular Component |
| mitochondrial alpha-ketoglutarate dehydrogenase complex |
| Function |
| catalytic activity |
| Molecular Function |
| 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity |
| alpha-ketoacid dehydrogenase activity |
| carboxy-lyase activity |
| Process |
| metabolic process |
|
| Cellular Location
|
- Mitochondrion matrix
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Valine, Leucine and Isoleucine Degradation |  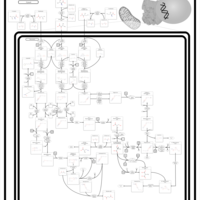 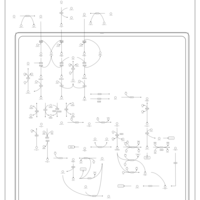 |  | | Beta-Ketothiolase Deficiency | 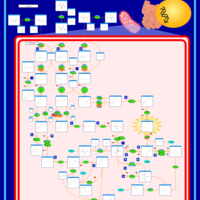 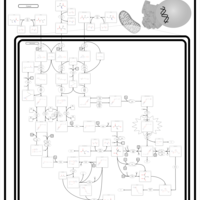 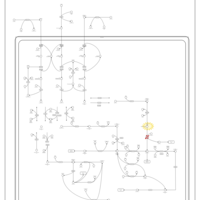 | Not Available | | 2-Methyl-3-Hydroxybutryl CoA Dehydrogenase Deficiency |  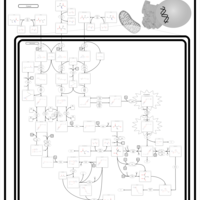 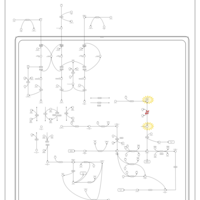 | Not Available | | Propionic Acidemia | 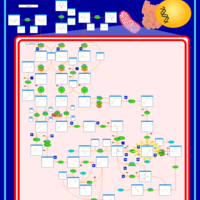 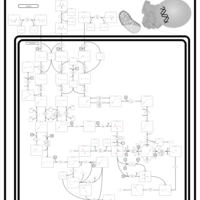 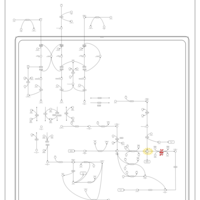 | Not Available | | 3-Hydroxy-3-Methylglutaryl-CoA Lyase Deficiency | 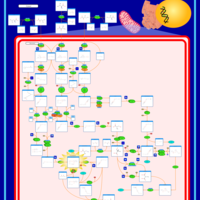 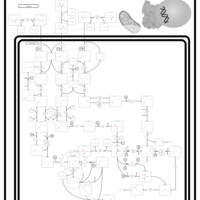 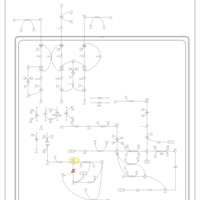 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 6 |
| Locus
| 6q14.1 |
| SNPs
| BCKDHB |
| Gene Sequence
|
>1179 bp
ATGGCGGTTGTAGCGGCGGCTGCCGGCTGGCTACTCAGGCTCAGGGCGGCAGGGGCTGAG
GGGCACTGGCGTCGGCTTCCTGGCGCGGGGCTGGCGCGGGGCTTTTTGCACCCCGCCGCG
ACTGTCGAGGATGCGGCCCAGAGGCGGCAGGTGGCTCATTTTACTTTCCAGCCAGATCCG
GAGCCCCGGGAGTACGGGCAAACTCAGAAAATGAATCTTTTCCAGTCTGTAACAAGTGCC
TTGGATAACTCATTGGCCAAAGATCCTACTGCAGTAATATTTGGTGAAGATGTTGCCTTT
GGTGGAGTCTTTAGATGCACTGTTGGCTTGCGAGACAAATATGGAAAAGATAGAGTTTTT
AATACCCCATTGTGTGAACAAGGAATTGTTGGATTTGGAATCGGAATTGCGGTCACTGGA
GCTACTGCCATTGCGGAAATTCAGTTTGCAGATTATATTTTCCCTGCATTTGATCAGATT
GTTAATGAAGCTGCCAAGTATCGCTATCGCTCTGGGGATCTTTTTAACTGTGGAAGCCTC
ACTATCCGGTCCCCTTGGGGCTGTGTTGGTCATGGGGCTCTCTATCATTCTCAGAGTCCT
GAAGCATTTTTTGCCCATTGCCCAGGAATCAAGGTGGTTATACCCAGAAGCCCTTTCCAG
GCCAAAGGACTTCTTTTGTCATGCATAGAGGATAAAAATCCTTGTATATTTTTTGAACCT
AAAATACTTTACAGGGCAGCAGCGGAAGAAGTCCCTATAGAACCATACAACATCCCACTG
TCCCAGGCCGAAGTCATACAGGAAGGGAGTGATGTTACTCTAGTTGCCTGGGGCACTCAG
GTTCATGTGATCCGAGAGGTAGCTTCCATGGCAAAAGAAAAGCTTGGAGTGTCTTGTGAA
GTCATTGATCTGAGGACTATAATACCTTGGGATGTGGACACAATTTGTAAGTCTGTGATC
AAAACAGGGCGACTGCTAATCAGTCACGAGGCTCCCTTGACAGGCGGCTTTGCATCGGAA
ATCAGCTCTACAGTTCAGGAGGAATGTTTCTTGAACCTAGAGGCTCCTATATCAAGAGTA
TGTGGTTATGACACACCATTTCCTCACATTTTTGAACCATTCTACATCCCAGACAAATGG
AAGTGTTATGATGCCCTTCGAAAAATGATCAACTATTGA
|
| Protein Properties |
|---|
| Number of Residues
| 392 |
| Molecular Weight
| 43122.065 |
| Theoretical pI
| 6.293 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>2-oxoisovalerate dehydrogenase subunit beta, mitochondrial
MAVVAAAAGWLLRLRAAGAEGHWRRLPGAGLARGFLHPAATVEDAAQRRQVAHFTFQPDP
EPREYGQTQKMNLFQSVTSALDNSLAKDPTAVIFGEDVAFGGVFRCTVGLRDKYGKDRVF
NTPLCEQGIVGFGIGIAVTGATAIAEIQFADYIFPAFDQIVNEAAKYRYRSGDLFNCGSL
TIRSPWGCVGHGALYHSQSPEAFFAHCPGIKVVIPRSPFQAKGLLLSCIEDKNPCIFFEP
KILYRAAAEEVPIEPYNIPLSQAEVIQEGSDVTLVAWGTQVHVIREVASMAKEKLGVSCE
VIDLRTIIPWDVDTICKSVIKTGRLLISHEAPLTGGFASEISSTVQEECFLNLEAPISRV
CGYDTPFPHIFEPFYIPDKWKCYDALRKMINY
|
| External Links |
|---|
| GenBank ID Protein
| 179362 |
| UniProtKB/Swiss-Prot ID
| P21953 |
| UniProtKB/Swiss-Prot Entry Name
| ODBB_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| M55575 |
| GeneCard ID
| BCKDHB |
| GenAtlas ID
| BCKDHB |
| HGNC ID
| HGNC:987 |
| References |
|---|
| General References
| Not Available |
