| Identification |
|---|
| HMDB Protein ID
| CDBP00430 |
| Secondary Accession Numbers
| Not Available |
| Name
| 2-oxoglutarate dehydrogenase, mitochondrial |
| Description
| Not Available |
| Synonyms
|
- 2-oxoglutarate dehydrogenase complex component E1
- Alpha-ketoglutarate dehydrogenase
- OGDC-E1
|
| Gene Name
| OGDH |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in oxoglutarate dehydrogenase (succinyl-transferring) activity |
| Specific Function
| The 2-oxoglutarate dehydrogenase complex catalyzes the overall conversion of 2-oxoglutarate to succinyl-CoA and CO(2). It contains multiple copies of three enzymatic components: 2-oxoglutarate dehydrogenase (E1), dihydrolipoamide succinyltransferase (E2) and lipoamide dehydrogenase (E3).
|
| GO Classification
|
| Biological Process |
| cellular nitrogen compound metabolic process |
| hippocampus development |
| olfactory bulb mitral cell layer development |
| tangential migration from the subventricular zone to the olfactory bulb |
| thalamus development |
| glycolysis |
| lysine catabolic process |
| tricarboxylic acid cycle |
| cerebellar cortex development |
| pyramidal neuron development |
| striatum development |
| Cellular Component |
| mitochondrial membrane |
| mitochondrial matrix |
| Function |
| oxidoreductase activity |
| thiamin pyrophosphate binding |
| oxoglutarate dehydrogenase (succinyl-transferring) activity |
| oxidoreductase activity, acting on the aldehyde or oxo group of donors |
| oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor |
| binding |
| catalytic activity |
| vitamin binding |
| Molecular Function |
| oxoglutarate dehydrogenase (NAD+) activity |
| oxoglutarate dehydrogenase (succinyl-transferring) activity |
| thiamine pyrophosphate binding |
| Process |
| metabolic process |
| small molecule metabolic process |
| alcohol metabolic process |
| monosaccharide metabolic process |
| hexose metabolic process |
| glucose metabolic process |
| glucose catabolic process |
| glycolysis |
|
| Cellular Location
|
- Mitochondrion matrix
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Citric Acid Cycle | 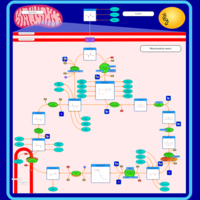 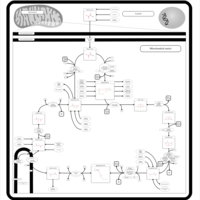 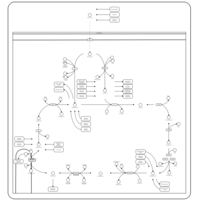 |  | | Lysine Degradation | 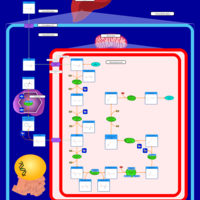 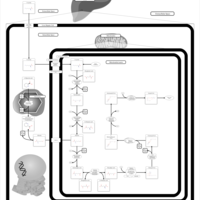 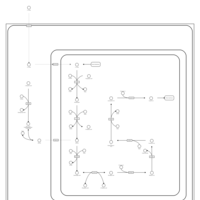 |  | | Citrate cycle (TCA cycle) | Not Available |  | | Congenital lactic acidosis | 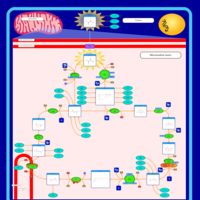 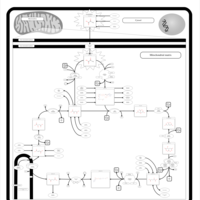 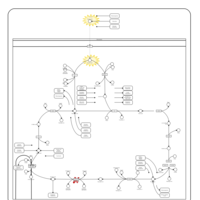 | Not Available | | Fumarase deficiency | 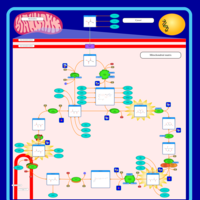 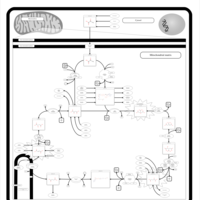 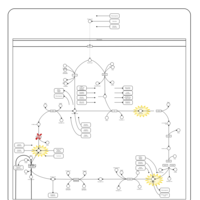 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 7 |
| Locus
| 7p14-p13 |
| SNPs
| OGDH |
| Gene Sequence
|
>3072 bp
ATGTTTCATTTAAGGACTTGTGCTGCTAAGTTGAGGCCATTGACGGCTTCCCAGACTGTT
AAGACATTTTCACAAAACAGACCAGCAGCAGCTAGGACATTTCAACAGATTCGGTGCTAT
TCTGCACCTGTTGCTGCTGAGCCCTTTCTCAGTGGGACTAGTTCGAACTATGTGGAGGAG
ATGTACTGTGCTTGGCTGGAAAACCCCAAAAGTGTACATAAGTCATGGGACATTTTTTTT
CGCAACACGAATGCCGGAGCCCCACCGGGCACTGCCTACCAGAGTCCCCTTCCCCTGAGC
CGAGGCTCCCTGGCTGCTGTGGCCCATGCACAGTCCCTGGTAGAAGCACAGCCCAACGTG
GACAAGCTCGTGGAGGACCACCTGGCAGTGCAGTCGCTCATCAGGGCATATCAGATACGA
GGGCACCATGTAGCACAGCTGGACCCCCTGGGGATTTTGGATGCTGATCTGGACTCCTCC
GTGCCCGCTGACATTATCTCATCCACAGACAAACTTGGGTTCTATGGCCTGGATGAGTCT
GACCTCGACAAGGTCTTCCACTTGCCCACCACCACTTTCATCGGGGGACAGGAATCAGCA
CTTCCTCTGCGGGAGATCATCCGTCGGCTGGAGATGGCCTACTGCCAGCATATTGGGGTG
GAGTTCATGTTCATCAATGACCTGGAGCAGTGCCAGTGGATCCGGCAGAAGTTTGAGACC
CCTGGGATCATGCAGTTCACAAATGAGGAGAAACGGACCCTGCTGGCCAGGCTTGTGCGG
TCCACCAGGTTTGAGGAGTTCCTACAGCGGAAGTGGTCCTCTGAGAAGCGCTTTGGTCTA
GAAGGCTGCGAGGTACTGATCCCTGCCCTCAAGACCATCATTGACAAGTCTAGTGAGAAT
GGCGTGGACTACGTGATCATGGGCATGCCACACAGAGGGCGGCTGAACGTGCTTGCAAAT
GTCATCAGGAAGGAGCTGGAACAGATCTTCTGTCAATTCGATTCAAAGCTGGAGGCAGCT
GATGAGGGCTCCGGAGATGTGAAGTACCACCTGGGCATGTATCACCGCAGGATCAATCGT
GTCACCGACAGGAACATTACCTTGTCCTTGGTGGCCAACCCTTCCCACCTTGAGGCCGCT
GACCCCGTGGTGATGGGCAAGACCAAAGCCGAACAGTTTTACTGTGGCGACACTGAAGGG
AAAAAGGTCATGTCCATCCTGTTGCATGGGGATGCTGCATTTGCTGGCCAGGGCATTGTG
TACGAGACCTTCCACCTCAGCGACCTGCCATCCTACACAACTCATGGCACCGTGCACGTG
GTCGTCAACAACCAGATCGGCTTCACCACCGACCCTCGGATGGCCCGCTCCTCCCCCTAC
CCCACTGACGTGGCCCGAGTGGTGAATGCCCCCATTTTCCACGTGAACTCAGATGACCCC
GAGGCTGTCATGTACGTGTGCAAAGTGGCGGCCGAGTGGAGGAGCACCTTCCACAAGGAC
GTGGTTGTCGATTTGGTGTGTTACCGGCGCAACGGCCACAACGAGATGGATGAGCCCATG
TTCACGCAGCCGCTCATGTACAAGCAGATCCGCAAGCAGAAGCCTGTGTTACAGAAGTAC
GCTGAGCTGCTGGTGTCGCAGGGTGTGGTCAACCAGCCTGAGTATGAGGAGGAAATTTCC
AAGTATGATAAGATCTGTGAGGAAGCTTTTGCCAGATCTAAAGATGAGAAGATCTTGCAC
ATTAAGCACTGGCTGGACTCTCCCTGGCCTGGCTTCTTCACCCTGGACGGGCAGCCCAGG
AGCATGTCCTGCCCCTCCACGGGTCTGACGGAGGATATTCTGACACACATCGGGAATGTG
GCTAGTTCTGTGCCTGTGGAAAACTTTACTATTCATGGAGGGCTGAGCCGGATCTTGAAG
ACTCGTGGGGAAATGGTGAAGAACCGGACTGTGGACTGGGCTCTAGCGGAGTACATGGCG
TTTGGCTCGCTCCTGAAGGAGGGCATCCACATTCGGCTGAGCGGCCAGGACGTGGAGCGG
GGCACATTCAGCCACCGCCACCATGTGCTCCATGACCAGAATGTGGACAAGAGAACCTGC
ATCCCCATGAACCATCTCTGGCCCAATCAGGCCCCCTATACTGTGTGCAACAGCTCACTG
TCTGAGTACGGCGTGCTGGGCTTTGAGCTGGGCTTCGCCATGGCCAGTCCTAATGCCCTG
GTCCTCTGGGAAGCCCAATTTGGTGACTTCCACAACACGGCCCAGTGTATCATCGACCAG
TTCATCTGCCCGGGACAAGCCAAGTGGGTGCGGCAGAATGGCATCGTGTTGCTGCTGCCC
CATGGCATGGAGGGCATGGGTCCAGAACATTCCTCCGCCCGCCCAGAGCGGTTCTTGCAG
ATGTGCAACGATGACCCAGATGTCCTGCCAGACCTTAAAGAAGCCAACTTCGACATCAAT
CAGCTATATGACTGCAATTGGGTTGTTGTCAACTGCTCCACTCCTGGCAACTTCTTCCAC
GTGCTACGACGCCAGATCCTGCTGCCATTCCGGAAGCCGTTAATTATCTTCACCCCCAAA
TCCCTGTTGCGCCACCCCGAGGCCAGATCCAGCTTTGATGAGATGCTTCCAGGAACCCAC
TTCCAGCGGGTGATCCCAGAAGATGGCCCTGCAGCTCAGAACCCAGAAAATGTCAAAAGG
CTTCTCTTCTGCACCGGCAAAGTGTATTATGACCTCACCCGGGAGCGCAAAGCACGCGAC
ATGGTGGGGCAGGTGGCCATCACAAGGATTGAGCAGCTGTCGCCATTCCCCTTTGACCTC
CTGCTGAAGGAGGTGCAGAAGTACCCCAATGCTGAGCTGGCCTGGTGCCAGGAGGAGCAC
AAGAACCAAGGCTACTATGACTACGTGAAGCCAAGACTTCGGACCACCATCAGCCGCGCC
AAGCCCGTCTGGTATGCCGGCCGGGACCCAGCGGCTGCTCCAGCCACCGGCAACAAGAAG
ACCCACCTGACGGAGCTGCAGCGCCTCCTGGACACGGCCTTCGACCTGGACGTCTTCAAG
AACTTCTCGTAG
|
| Protein Properties |
|---|
| Number of Residues
| 1023 |
| Molecular Weight
| 48179.59 |
| Theoretical pI
| 7.895 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>2-oxoglutarate dehydrogenase, mitochondrial
MFHLRTCAAKLRPLTASQTVKTFSQNRPAAARTFQQIRCYSAPVAAEPFLSGTSSNYVEE
MYCAWLENPKSVHKSWDIFFRNTNAGAPPGTAYQSPLPLSRGSLAAVAHAQSLVEAQPNV
DKLVEDHLAVQSLIRAYQIRGHHVAQLDPLGILDADLDSSVPADIISSTDKLGFYGLDES
DLDKVFHLPTTTFIGGQESALPLREIIRRLEMAYCQHIGVEFMFINDLEQCQWIRQKFET
PGIMQFTNEEKRTLLARLVRSTRFEEFLQRKWSSEKRFGLEGCEVLIPALKTIIDKSSEN
GVDYVIMGMPHRGRLNVLANVIRKELEQIFCQFDSKLEAADEGSGDVKYHLGMYHRRINR
VTDRNITLSLVANPSHLEAADPVVMGKTKAEQFYCGDTEGKKVMSILLHGDAAFAGQGIV
YETFHLSDLPSYTTHGTVHVVVNNQIGFTTDPRMARSSPYPTDVARVVNAPIFHVNSDDP
EAVMYVCKVAAEWRSTFHKDVVVDLVCYRRNGHNEMDEPMFTQPLMYKQIRKQKPVLQKY
AELLVSQGVVNQPEYEEEISKYDKICEEAFARSKDEKILHIKHWLDSPWPGFFTLDGQPR
SMSCPSTGLTEDILTHIGNVASSVPVENFTIHGGLSRILKTRGEMVKNRTVDWALAEYMA
FGSLLKEGIHIRLSGQDVERGTFSHRHHVLHDQNVDKRTCIPMNHLWPNQAPYTVCNSSL
SEYGVLGFELGFAMASPNALVLWEAQFGDFHNTAQCIIDQFICPGQAKWVRQNGIVLLLP
HGMEGMGPEHSSARPERFLQMCNDDPDVLPDLKEANFDINQLYDCNWVVVNCSTPGNFFH
VLRRQILLPFRKPLIIFTPKSLLRHPEARSSFDEMLPGTHFQRVIPEDGPAAQNPENVKR
LLFCTGKVYYDLTRERKARDMVGQVAITRIEQLSPFPFDLLLKEVQKYPNAELAWCQEEH
KNQGYYDYVKPRLRTTISRAKPVWYAGRDPAAAPATGNKKTHLTELQRLLDTAFDLDVFK
NFS
|
| External Links |
|---|
| GenBank ID Protein
| 37674435 |
| UniProtKB/Swiss-Prot ID
| Q02218 |
| UniProtKB/Swiss-Prot Entry Name
| ODO1_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AC004859 |
| GeneCard ID
| OGDH |
| GenAtlas ID
| OGDH |
| HGNC ID
| HGNC:8124 |
| References |
|---|
| General References
| Not Available |

