| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:20:04 UTC |
|---|
| Updated at | 2020-12-07 19:11:46 UTC |
|---|
| CannabisDB ID | CDB005207 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Inositol 1,4,5-trisphosphate |
|---|
| Description | Inositol 1,4,5-trisphosphate, also known as 1,4,5-insp3 or IP3, belongs to the class of organic compounds known as inositol phosphates. Inositol phosphates are compounds containing a phosphate group attached to an inositol (or cyclohexanehexol) moiety. Inositol 1,4,5-trisphosphate is an extremely weak basic (essentially neutral) compound (based on its pKa). Inositol 1,4,5-trisphosphate exists in all eukaryotes, ranging from yeast to humans. DG(16:0/18:1(9Z)/0:0) and inositol 1,4,5-trisphosphate can be biosynthesized from pip2[4,5](16:0/18:1(9Z)); which is catalyzed by the enzyme 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1. In humans, inositol 1,4,5-trisphosphate is involved in the metabolic disorder called the joubert syndrome pathway. Inositol 1,4,5-trisphosphate is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,4,5-Insp3 | ChEBI | | D-Myo-inositol 1,4,5-trisphosphate | ChEBI | | D-MYO-inositol-1,4,5-triphosphATE | ChEBI | | Ins(1,4,5)P3 | ChEBI | | InsP3 | ChEBI | | IP3 | ChEBI | | 1D-Myo-inositol 1,4,5-trisphosphate | Kegg | | D-Myo-inositol 1,4,5-trisphosphoric acid | Generator | | D-MYO-inositol-1,4,5-triphosphoric acid | Generator | | 1D-Myo-inositol 1,4,5-trisphosphoric acid | Generator | | Inositol 1,4,5-trisphosphoric acid | Generator | | Myo-inositol 1,4,5-trisphosphate | HMDB | | 1,4,5-IP3 | HMDB | | Inositol 1,4,5-triphosphate | HMDB | | Myoinositol 1,4,5-triphosphate | HMDB | | D-myo-Inositol 1,4,5-triphosphate | HMDB | | IP 3 | HMDB | | Inositol 1,4,5-trisphosphate | HMDB | | Inositol triphosphate | HMDB | | Inositol trisphosphate | HMDB | | Triphosphoinositol | HMDB | | myo-Inositol 1,4,5-triphosphate | HMDB | | myo-Inositol triphosphate | HMDB | | myo-Inositol trisphosphate | HMDB |
|
|---|
| Chemical Formula | C6H15O15P3 |
|---|
| Average Molecular Weight | 420.1 |
|---|
| Monoisotopic Molecular Weight | 419.9624 |
|---|
| IUPAC Name | {[(1R,2S,3R,4R,5S,6R)-2,3,5-trihydroxy-4,6-bis(phosphonooxy)cyclohexyl]oxy}phosphonic acid |
|---|
| Traditional Name | InsP3 |
|---|
| CAS Registry Number | 85166-31-0 |
|---|
| SMILES | O[C@@H]1[C@H](O)[C@@H](OP(O)(O)=O)[C@H](OP(O)(O)=O)[C@@H](O)[C@@H]1OP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H15O15P3/c7-1-2(8)5(20-23(13,14)15)6(21-24(16,17)18)3(9)4(1)19-22(10,11)12/h1-9H,(H2,10,11,12)(H2,13,14,15)(H2,16,17,18)/t1-,2+,3+,4-,5-,6-/m1/s1 |
|---|
| InChI Key | MMWCIQZXVOZEGG-XJTPDSDZSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as inositol phosphates. Inositol phosphates are compounds containing a phosphate group attached to an inositol (or cyclohexanehexol) moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Alcohols and polyols |
|---|
| Direct Parent | Inositol phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Inositol phosphate
- Monoalkyl phosphate
- Cyclohexanol
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Secondary alcohol
- Polyol
- Organic oxide
- Hydrocarbon derivative
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Inositol 1,4,5-trisphosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-002b-7195200000-2871156f7c1e00704088 | Spectrum | | Predicted GC-MS | Inositol 1,4,5-trisphosphate, 3 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0002-9052133000-5094b6ef55759c5a19dd | Spectrum | | Predicted GC-MS | Inositol 1,4,5-trisphosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-0006-0091000000-66258d5943e8e34a4f1b | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-00ba-9030000000-9567cb0f36d067bd98e8 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 13V, negative | splash10-0udi-0000900000-a08362b50609e7868113 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 19V, negative | splash10-0udi-0205900000-c9a653bc0177b1f5ec0c | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 20V, negative | splash10-0udi-1509800000-0c58a55629c8bdde91c4 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 25V, negative | splash10-0zi0-2709200000-f834442aa900d5925ee0 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 29V, negative | splash10-0kk9-6908000000-bce10d89581b945182c8 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 34V, negative | splash10-056r-9603000000-b83a25f979d90dcaacec | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 40V, negative | splash10-004i-9300000000-83996fe12a20b170e2dc | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 47V, negative | splash10-004i-9100000000-9c6fa50af864d3ab9ccc | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-0udi-0109000000-f750488015579ddac395 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-004i-9000000000-0ae8d2338f48c17984cd | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-0zfr-0970000000-d129d480332834af8631 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-004r-5900000000-4dffa7896e57b68d360e | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-08fr-0639000000-1fc55e97bbb62fd98506 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 29V, negative | splash10-0a4i-0943000000-4b6820f7da0e5d559167 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 13V, negative | splash10-014i-0000900000-4422ae3f5c5cbce0813d | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 16V, negative | splash10-014i-0103900000-60bb04a7db68b52b301a | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 22V, negative | splash10-05fr-1409300000-c28e47a55145868de89d | 2020-07-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-3008900000-e987cfb2603d94f1a6a4 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0uk9-2006900000-95047338604f8928313f | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-4291000000-8e9bb388f01ec6d535ab | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-4001900000-395b5e207aa26dd43f27 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9003200000-5e6369906fdedd08ad60 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-4b72b937fe3bf7fe817e | 2016-09-12 | View Spectrum |
|
|---|
| NMR | Not Available |
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Inositol Metabolism | 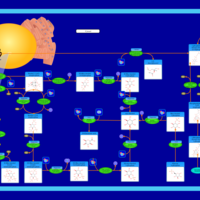 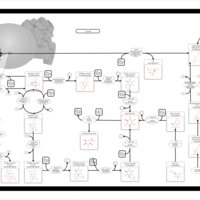 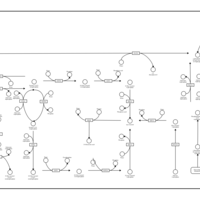 |  | | Phosphatidylinositol Phosphate Metabolism | 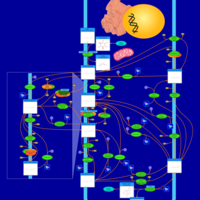 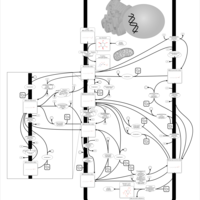 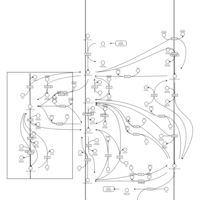 |  | | Inositol Phosphate Metabolism | 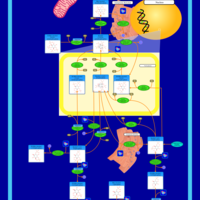 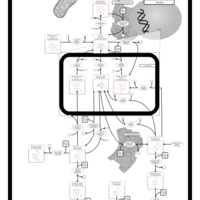 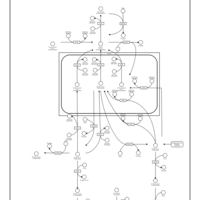 |  | | Joubert syndrome | 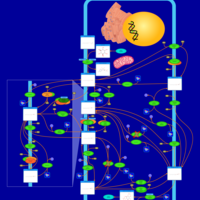 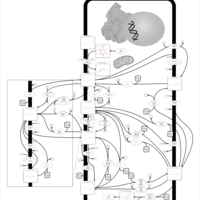 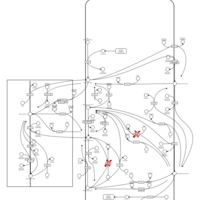 | Not Available | | Muscle/Heart Contraction | 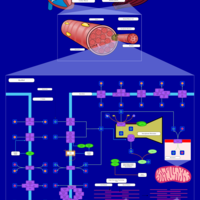 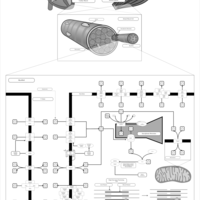  | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | PLCB1 | 20p12 | Q9NQ66 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-4 | PLCB4 | 20p12 | Q15147 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-2 | PLCB2 | 15q15 | Q00722 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3 | PLCB3 | 11q13 | Q01970 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-2 | PLCG2 | 16q24.1 | P16885 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | PLCG1 | 20q12-q13.1 | P19174 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1 | PLCD1 | 3p22-p21.3 | P51178 | details | | Type I inositol 1,4,5-trisphosphate 5-phosphatase | INPP5A | 10q26.3 | Q14642 | details | | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A | INPP5J | 22q12.2 | Q15735 | details | | Inositol polyphosphate 5-phosphatase K | INPP5K | 17p13.3 | Q9BT40 | details | | Inositol-trisphosphate 3-kinase B | ITPKB | 1q42.13 | P27987 | details | | Inositol-trisphosphate 3-kinase A | ITPKA | 15q15.1 | P23677 | details | | Inositol polyphosphate multikinase | IPMK | 10q21.1 | Q8NFU5 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase eta-2 | PLCH2 | 1p36.32 | O75038 | details | | Inositol-trisphosphate 3-kinase C | ITPKC | 19q13.1 | Q96DU7 | details | | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | INPPL1 | 11q13 | O15357 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-4 | PLCD4 | 2q35 | Q9BRC7 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase eta-1 | PLCH1 | 3q25.31 | Q4KWH8 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase epsilon-1 | PLCE1 | 10q23 | Q9P212 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-3 | PLCD3 | 17q21.31 | Q8N3E9 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 | PLCZ1 | 12p12.3 | Q86YW0 | details | | Multiple inositol polyphosphate phosphatase 1 | MINPP1 | 10q23 | Q9UNW1 | details |
|
|---|
| Transporters | |
| Inositol 1,4,5-trisphosphate receptor type 1 | ITPR1 | 3p26.1 | Q14643 | details | | Inositol 1,4,5-trisphosphate receptor type 2 | ITPR2 | 12p11 | Q14571 | details | | Inositol 1,4,5-trisphosphate receptor type 3 | ITPR3 | 6p21 | Q14573 | details |
|
|---|
| Metal Bindings | |
| 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | PLCB1 | 20p12 | Q9NQ66 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-4 | PLCB4 | 20p12 | Q15147 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-2 | PLCB2 | 15q15 | Q00722 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3 | PLCB3 | 11q13 | Q01970 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1 | PLCD1 | 3p22-p21.3 | P51178 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase eta-2 | PLCH2 | 1p36.32 | O75038 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-4 | PLCD4 | 2q35 | Q9BRC7 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase eta-1 | PLCH1 | 3q25.31 | Q4KWH8 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase epsilon-1 | PLCE1 | 10q23 | Q9P212 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-3 | PLCD3 | 17q21.31 | Q8N3E9 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 | PLCZ1 | 12p12.3 | Q86YW0 | details |
|
|---|
| Receptors | |
| 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | PLCB1 | 20p12 | Q9NQ66 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3 | PLCB3 | 11q13 | Q01970 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-2 | PLCG2 | 16q24.1 | P16885 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | PLCG1 | 20q12-q13.1 | P19174 | details | | Inositol polyphosphate 5-phosphatase K | INPP5K | 17p13.3 | Q9BT40 | details | | Inositol-trisphosphate 3-kinase B | ITPKB | 1q42.13 | P27987 | details | | Inositol 1,4,5-trisphosphate receptor type 1 | ITPR1 | 3p26.1 | Q14643 | details | | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | INPPL1 | 11q13 | O15357 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase epsilon-1 | PLCE1 | 10q23 | Q9P212 | details | | Inositol 1,4,5-trisphosphate receptor type 2 | ITPR2 | 12p11 | Q14571 | details | | Inositol 1,4,5-trisphosphate receptor type 3 | ITPR3 | 6p21 | Q14573 | details |
|
|---|
| Transcriptional Factors | |
| 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | PLCB1 | 20p12 | Q9NQ66 | details | | Inositol polyphosphate 5-phosphatase K | INPP5K | 17p13.3 | Q9BT40 | details |
|
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0001498 |
|---|
| DrugBank ID | DB03401 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB022657 |
|---|
| KNApSAcK ID | C00007460 |
|---|
| Chemspider ID | 388562 |
|---|
| KEGG Compound ID | C01245 |
|---|
| BioCyc ID | INOSITOL-1-4-5-TRISPHOSPHATE |
|---|
| BiGG ID | 1485108 |
|---|
| Wikipedia Link | Inositol trisphosphate |
|---|
| METLIN ID | 6282 |
|---|
| PubChem Compound | 439456 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16595 |
|---|
| References |
|---|
| General References | Not Available |
|---|
