| Identification |
|---|
| HMDB Protein ID
| CDBP02581 |
| Secondary Accession Numbers
| Not Available |
| Name
| 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 |
| Description
| Not Available |
| Synonyms
|
- PLC-zeta-1
- Phosphoinositide phospholipase C-zeta-1
- Phospholipase C-zeta-1
- Testis-development protein NYD-SP27
|
| Gene Name
| PLCZ1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in calcium ion binding |
| Specific Function
| The production of the second messenger molecules diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3) is mediated by activated phosphatidylinositol-specific phospholipase C enzymes. In vitro, hydrolyzes PtdIns(4,5)P2 in a Ca(2+)-dependent manner. Triggers intracellular Ca(2+) oscillations in oocytes solely during M phase and is involved in inducing oocyte activation and initiating embryonic development up to the blastocyst stage. Is therefore a strong candidate for the egg-activating soluble sperm factor that is transferred from the sperm into the egg cytoplasm following gamete membrane fusion. May exert an inhibitory effect on phospholipase-C-coupled processes that depend on calcium ions and protein kinase C, including CFTR trafficking and function.
|
| GO Classification
|
| Biological Process |
| multicellular organismal development |
| lipid catabolic process |
| egg activation |
| calcium ion transport |
| intracellular signal transduction |
| Cellular Component |
| perinuclear region of cytoplasm |
| nucleus |
| Function |
| catalytic activity |
| hydrolase activity |
| phosphoric ester hydrolase activity |
| phosphoric diester hydrolase activity |
| phospholipase c activity |
| phosphoinositide phospholipase c activity |
| calcium ion binding |
| lipase activity |
| phospholipase activity |
| ion binding |
| cation binding |
| metal ion binding |
| hydrolase activity, acting on ester bonds |
| binding |
| Molecular Function |
| phosphatidylinositol phospholipase C activity |
| signal transducer activity |
| calcium ion binding |
| Process |
| regulation of cellular process |
| signal transduction |
| lipid metabolic process |
| intracellular signaling pathway |
| signaling |
| signaling pathway |
| metabolic process |
| primary metabolic process |
| biological regulation |
| regulation of biological process |
|
| Cellular Location
|
- Nucleus
- Cytoplasm
- perinuclear region
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Calcium signaling pathway | Not Available | 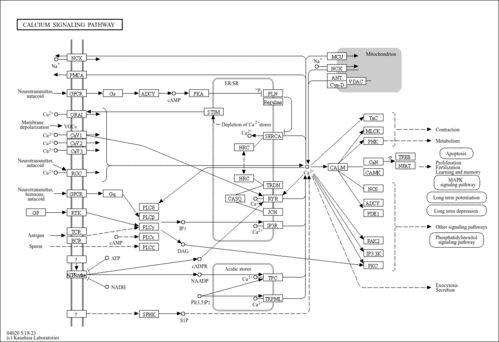 | | Phosphatidylinositol signaling system | Not Available | 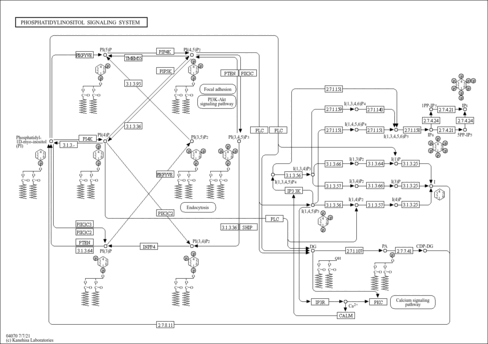 | | Oocyte meiosis | Not Available | 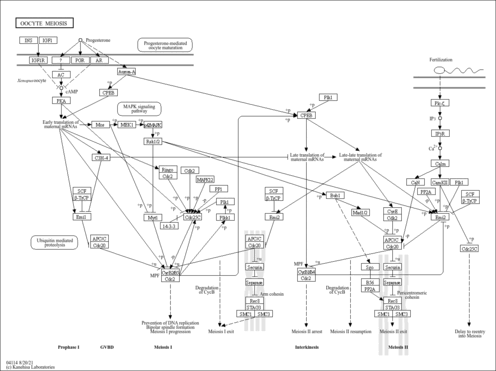 |
|
| Gene Properties |
|---|
| Chromosome Location
| 12 |
| Locus
| 12p12.3 |
| SNPs
| PLCZ1 |
| Gene Sequence
|
>1827 bp
ATGGAAATGAGATGGTTTTTGTCAAAGATTCAGGATGACTTCAGAGGTGGAAAAATTAAC
CTAGAAAAAACTCAGAGGTTACTTGAAAAATTAGATATTCGGTGCAGTTATATTCATGTG
AAACAGATTTTTAAGGACAATGACAGGCTGAAACAAGGAAGAATCACCATAGAAGAATTT
AGAGCAATTTATCGAATTATCACGCACAGAGAAGAAATTATTGAGATTTTCAACACATAT
TCTGAAAACCGGAAAATTCTTTTAGCAAGTAATCTGGCTCAATTTCTGACACAAGAACAA
TATGCAGCTGAGATGAGTAAAGCTATTGCTTTTGAGATCATTCAGAAATACGAGCCTATC
GAAGAAGTTAGGAAAGCACACCAAATGTCATTAGAAGGTTTTACAAGATACATGGATTCA
CGTGAATGTCTACTGTTTAAAAATGAATGTAGAAAAGTTTATCAAGATATGACTCATCCA
TTAAATGATTATTTTATTTCATCTTCACATAACACATATTTGGTATCTGATCAATTATTG
GGACCAAGTGACCTTTGGGGATATGTAAGTGCCCTTGTGAAAGGATGCCGTTGTTTGGAG
ATTGACTGCTGGGATGGAGCACAAAATGAACCTGTTGTATATCATGGCTACACACTCACA
AGCAAACTTCTGTTTAAAACTGTTATCCAAGCTATACACAAGTATGCATTCATGACATCT
GACTACCCAGTGGTGCTCTCTTTAGAAAATCACTGCTCCACTGCCCAACAAGAAGTAATG
GCAGACAATTTGCAGGCTACTTTTGGAGAGTCCTTGCTTTCTGATATGCTTGATGATTTT
CCTGATACTCTACCATCACCAGAGGCACTAAAATTCAAAATATTAGTTAAAAATAAGAAA
ATAGGAACCTTAAAGGAAACCCATGAAAGAAAAGGTTCTGATAAGCGTGGAGACAATCAA
GACAAGGAAACAGGGGTAAAAAAGTTACCTGGAGTAATGCTTTTCAAGAAAAAGAAGACC
AGGAAGCTAAAAATTGCTCTGGCCTTATCTGATCTTGTCATTTATACGAAAGCTGAGAAA
TTCAAAAGCTTTCAACATTCAAGATTATATCAGCAATTTAATGAAAATAATTCTATTGGG
GAGACACAAGCCCGAAAACTTTCAAAATTGCGAGTCCATGAGTTTATTTTTCACACCAGG
AAGTTCATTACCAGAATATATCCCAAAGCAACAAGAGCAGACTCTTCTAATTTTAATCCC
CAAGAATTTTGGAATATAGGTTGTCAAATGGTGGCTTTAAATTTCCAGACCCCTGGTCTG
CCCATGGATCTGCAAAATGGGAAATTTTTGGATAATGGTGGTTCTGGATATATTTTGAAA
CCACATTTCTTAAGAGAGAGTAAATCATACTTTAACCCAAGTAACATAAAAGAGGGTATG
CCAATTACACTTACAATAAGGCTCATCAGTGGTATCCAGTTGCCTCTTACTCATTCATCA
TCTAACAAAGGTGATTCATTAGTAATTATAGAAGTTTTTGGTGTTCCAAATGATCAAATG
AAGCAGCAGACTCGTGTAATTAAAAAAAATGCTTTTAGTCCAAGATGGAATGAAACATTC
ACATTTATTATTCATGTCCCAGAATTGGCATTGATACGTTTTGTTGTTGAAGGTCAAGGT
TTAATAGCAGGAAATGAATTTCTTGGGCAATATACTTTGCCACTTCTATGCATGAACAAA
GGTTATCGTCGTATTCCTCTGTTTTCCAGAATGGGTGAGAGCCTTGAGCCTGCTTCACTG
TTTGTTTATGTTTGGTACGTCAGATAA
|
| Protein Properties |
|---|
| Number of Residues
| 608 |
| Molecular Weight
| 70410.725 |
| Theoretical pI
| 9.034 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase zeta-1
MEMRWFLSKIQDDFRGGKINLEKTQRLLEKLDIRCSYIHVKQIFKDNDRLKQGRITIEEF
RAIYRIITHREEIIEIFNTYSENRKILLASNLAQFLTQEQYAAEMSKAIAFEIIQKYEPI
EEVRKAHQMSLEGFTRYMDSRECLLFKNECRKVYQDMTHPLNDYFISSSHNTYLVSDQLL
GPSDLWGYVSALVKGCRCLEIDCWDGAQNEPVVYHGYTLTSKLLFKTVIQAIHKYAFMTS
DYPVVLSLENHCSTAQQEVMADNLQATFGESLLSDMLDDFPDTLPSPEALKFKILVKNKK
IGTLKETHERKGSDKRGDNQDKETGVKKLPGVMLFKKKKTRKLKIALALSDLVIYTKAEK
FKSFQHSRLYQQFNENNSIGETQARKLSKLRVHEFIFHTRKFITRIYPKATRADSSNFNP
QEFWNIGCQMVALNFQTPGLPMDLQNGKFLDNGGSGYILKPHFLRESKSYFNPSNIKEGM
PITLTIRLISGIQLPLTHSSSNKGDSLVIIEVFGVPNDQMKQQTRVIKKNAFSPRWNETF
TFIIHVPELALIRFVVEGQGLIAGNEFLGQYTLPLLCMNKGYRRIPLFSRMGESLEPASL
FVYVWYVR
|
| External Links |
|---|
| GenBank ID Protein
| 25053795 |
| UniProtKB/Swiss-Prot ID
| Q86YW0 |
| UniProtKB/Swiss-Prot Entry Name
| PLCZ1_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| AF532185 |
| GeneCard ID
| PLCZ1 |
| GenAtlas ID
| PLCZ1 |
| HGNC ID
| HGNC:19218 |
| References |
|---|
| General References
| Not Available |


