| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:50:16 UTC |
|---|
| Updated at | 2020-12-07 19:11:16 UTC |
|---|
| CannabisDB ID | CDB004917 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | 3-Dehydrosphinganine |
|---|
| Description | 3-Dehydrosphinganine, also known as KDHS or ketodihydrosphingosine, belongs to the class of organic compounds known as beta-hydroxy ketones. These are ketones containing a hydroxyl group attached to the beta-carbon atom, relative to the C=O group. Thus, 3-dehydrosphinganine is considered to be a sphingoid base lipid molecule. 3-Dehydrosphinganine is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. 3-Dehydrosphinganine exists in all eukaryotes, ranging from yeast to humans. Within humans, 3-dehydrosphinganine participates in a number of enzymatic reactions. In particular, 3-dehydrosphinganine and carbon dioxide can be biosynthesized from palmityl-CoA and L-serine through the action of the enzyme serine palmitoyltransferase 1. In addition, 3-dehydrosphinganine can be biosynthesized from sphinganine; which is catalyzed by the enzyme 3-ketodihydrosphingosine reductase. In humans, 3-dehydrosphinganine is involved in the metabolic disorder called the krabbe disease pathway. Outside of the human body, 3-Dehydrosphinganine has been detected, but not quantified in, several different foods, such as common chokecherries, black huckleberries, pomegranates, cinnamons, and flaxseeds. This could make 3-dehydrosphinganine a potential biomarker for the consumption of these foods. A 2-amino-1-hydroxyoctadecan-3-one that has S-configuration. 3-Dehydrosphinganine is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3-Dehydro-D-sphinganine | ChEBI | | 3-Ketodihydrosphingosine | ChEBI | | 3-Ketosphinganine | ChEBI | | (2S)-2-Amino-1-hydroxyoctadecan-3-one | HMDB | | 1-Hydroxy-2-amino-3-oxo-octadecane | HMDB | | 2-Amino-1-hydroxy-3-octadecanone | HMDB | | KDHS | HMDB | | Ketodihydrosphingosine | HMDB | | (+-)-Isomer OF ketodihydrosphingosine | HMDB | | (S)-Isomer OF ketodihydrosphingosine | HMDB |
|
|---|
| Chemical Formula | C18H37NO2 |
|---|
| Average Molecular Weight | 299.49 |
|---|
| Monoisotopic Molecular Weight | 299.2824 |
|---|
| IUPAC Name | (2S)-2-amino-1-hydroxyoctadecan-3-one |
|---|
| Traditional Name | 3-ketosphinganine |
|---|
| CAS Registry Number | 16105-69-4 |
|---|
| SMILES | CCCCCCCCCCCCCCCC(=O)[C@@H](N)CO |
|---|
| InChI Identifier | InChI=1S/C18H37NO2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-18(21)17(19)16-20/h17,20H,2-16,19H2,1H3/t17-/m0/s1 |
|---|
| InChI Key | KBUNOSOGGAARKZ-KRWDZBQOSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as beta-hydroxy ketones. These are ketones containing a hydroxyl group attached to the beta-carbon atom, relative to the C=O group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbonyl compounds |
|---|
| Direct Parent | Beta-hydroxy ketones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Beta-hydroxy ketone
- Alpha-aminoketone
- Organic nitrogen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Primary amine
- Primary alcohol
- Organonitrogen compound
- Primary aliphatic amine
- Amine
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | 3-Dehydrosphinganine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-01p9-9120000000-135052b999433b5dd30b | Spectrum | | Predicted GC-MS | 3-Dehydrosphinganine, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-03di-9321000000-e0d96cb3bea3c320412a | Spectrum | | Predicted GC-MS | 3-Dehydrosphinganine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | 3-Dehydrosphinganine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f89-2096000000-8323a157b44b31f2331a | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01qa-4491000000-39be7a6025f78d062cb5 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01ox-9510000000-a675e52903f069501f67 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-1090000000-e36f44a44812cc09023c | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-015j-4090000000-4a24200105a7fd0afd1e | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9020000000-edd2ec1e7a7fae113d2f | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ue9-3069000000-e43a689cbba7742dc5c6 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01q9-9282000000-f83e19a33602a8a25cb8 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-9000000000-f16a410d8c3330af51f2 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014j-0090000000-7d1c46d2460baf58b0dc | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-06ej-3090000000-b775508a0489f38a77f7 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9010000000-b10c9eb2adca17092c8c | 2021-09-22 | View Spectrum |
|
|---|
| NMR | Not Available |
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Sphingolipid Metabolism | 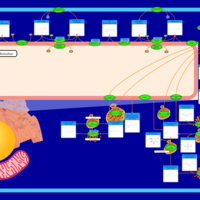 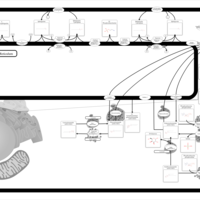 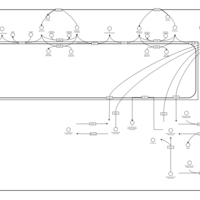 |  | | Gaucher Disease | 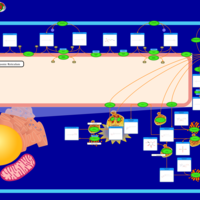 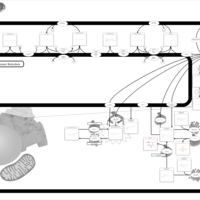 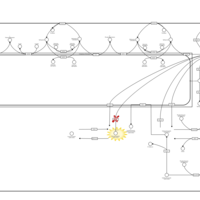 | Not Available | | Globoid Cell Leukodystrophy | 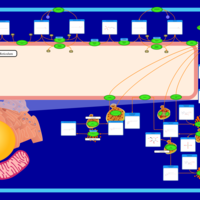 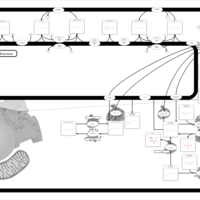 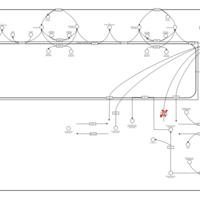 | Not Available | | Metachromatic Leukodystrophy (MLD) | 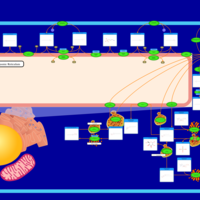 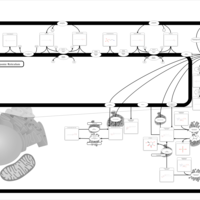 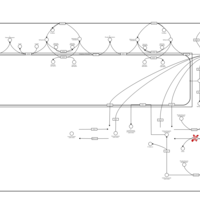 | Not Available | | Fabry disease | 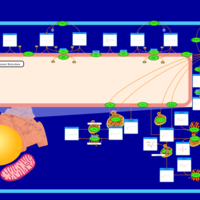 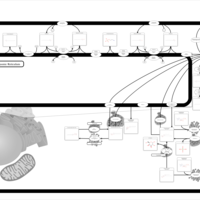 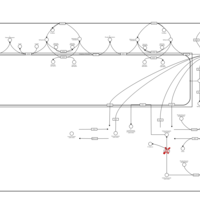 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Globoside alpha-1,3-N-acetylgalactosaminyltransferase 1 | GBGT1 | 9q34.13-q34.3 | Q8N5D6 | details | | N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase | PIGL | 17p12-p11.2 | Q9Y2B2 | details | | Serine palmitoyltransferase 1 | SPTLC1 | 9q22.2 | O15269 | details | | Serine palmitoyltransferase 2 | SPTLC2 | 14q24.3 | O15270 | details | | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q | PIGQ | 16p13.3 | Q9BRB3 | details | | Phosphatidylinositol N-acetylglucosaminyltransferase subunit A | PIGA | Xp22.1 | P37287 | details | | Phosphatidylinositol N-acetylglucosaminyltransferase subunit H | PIGH | 14q24.1 | Q14442 | details | | Phosphatidylinositol N-acetylglucosaminyltransferase subunit P | PIGP | 21q22.2 | P57054 | details | | Phosphatidylinositol N-acetylglucosaminyltransferase subunit C | PIGC | 1q23-q25 | Q92535 | details | | Ganglioside GM2 activator | GM2A | 5q33.1 | P17900 | details | | T-cell surface glycoprotein CD1e, membrane-associated | CD1E | 1q22-q23 | P15812 | details | | Epididymal secretory protein E1 | NPC2 | 14q24.3 | P61916 | details | | Antigen-presenting glycoprotein CD1d | CD1D | 1q22-q23 | P15813 | details | | GPI mannosyltransferase 1 | PIGM | 1q23.2 | Q9H3S5 | details | | Phosphatidylinositol-glycan biosynthesis class W protein | PIGW | | Q7Z7B1 | details | | 3-ketodihydrosphingosine reductase | KDSR | 18q21.3 | Q06136 | details | | Phosphatidylinositol-glycan biosynthesis class X protein | PIGX | 3q29 | Q8TBF5 | details | | GPI mannosyltransferase 4 | PIGZ | 3q29 | Q86VD9 | details | | Lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase | B3GNT5 | 3q28 | Q9BYG0 | details | | Beta-1,3-galactosyltransferase 5 | B3GALT5 | 21q22.3 | Q9Y2C3 | details | | GPI mannosyltransferase 3 | PIGB | 15q21-q22 | Q92521 | details | | Phosphatidylinositol-glycan biosynthesis class F protein | PIGF | 2p21-p16 | Q07326 | details | | GPI ethanolamine phosphate transferase 2 | PIGG | 4p16.3 | Q5H8A4 | details | | GPI ethanolamine phosphate transferase 1 | PIGN | 18q21.33 | O95427 | details | | GPI ethanolamine phosphate transferase 3 | PIGO | 9p13.3 | Q8TEQ8 | details | | GPI transamidase component PIG-S | PIGS | 17p13.2 | Q96S52 | details | | GPI transamidase component PIG-T | PIGT | 20q12-q13.12 | Q969N2 | details | | Phosphatidylinositol glycan anchor biosynthesis class U protein | PIGU | 20q11.22 | Q9H490 | details | | GPI mannosyltransferase 2 | PIGV | 1p36.11 | Q9NUD9 | details | | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Y | PIGY | 4q22.1 | Q3MUY2 | details | | Serine palmitoyltransferase 3 | SPTLC3 | 20p12.1 | Q9NUV7 | details | | Non-lysosomal glucosylceramidase | GBA2 | 9p13.3 | Q9HCG7 | details | | GPI-anchor transamidase | PIGK | 1p31.1 | Q92643 | details |
|
|---|
| Transporters | |
| Pleckstrin homology domain-containing family A member 8 | PLEKHA8 | 7p21-p11.2 | Q96JA3 | details | | Protein PLEKHA9 | PLEKHA9 | 12q | O95397 | details | | Glycolipid transfer protein domain-containing protein 2 | GLTPD2 | 17p13.2 | A6NH11 | details | | Putative uncharacterized protein PLEKHA8 | PLEKHA8 | 7p21-p11.2 | B5MDU3 | details | | Glycolipid transfer protein domain-containing protein 1 | GLTPD1 | 1p36.33 | Q5TA50 | details | | Putative uncharacterized protein DKFZp434L0435 | DKFZp434L0435 | | Q9UFH6 | details | | Glycolipid transfer protein | GLTP | 12q24.11 | Q9NZD2 | details |
|
|---|
| Metal Bindings | Not Available |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0001480 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB022645 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 388895 |
|---|
| KEGG Compound ID | C02934 |
|---|
| BioCyc ID | DEHYDROSPHINGANINE |
|---|
| BiGG ID | 1453300 |
|---|
| Wikipedia Link | Not Available |
|---|
| METLIN ID | 3428 |
|---|
| PubChem Compound | 439853 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17862 |
|---|
| References |
|---|
| General References | Not Available |
|---|
