| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:43:33 UTC |
|---|
| Updated at | 2022-12-13 23:36:24 UTC |
|---|
| CannabisDB ID | CDB004853 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | L-Glutamine |
|---|
| Description | L-Glutamine, also known as levoglutamide or Q, belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. An optically active form of glutamine having L-configuration. L-Glutamine is a drug which is used for nutritional supplementation, also for treating dietary shortage or imbalance.
used to reduce the acute complications of sickle cell disease in adult and pediatric patients 5 years of age and older [fda label]. L-Glutamine is a very strong basic compound (based on its pKa). L-Glutamine exists in all living species, ranging from bacteria to humans. phenylacetyl-CoA and L-glutamine can be converted into phenylacetylglutamine; which is mediated by the enzyme glycine N-acyltransferase. In humans, L-glutamine is involved in phenylacetate metabolism. Outside of the human body, L-Glutamine is found, on average, in the highest concentration within a few different foods, such as wheats, broccoli, and red beetroots and in a lower concentration in cucumbers, garden onions, and nanking cherries. L-Glutamine has also been detected, but not quantified in, several different foods, such as rambutans, common salsifies, greenthread tea, common walnuts, and celery stalks. This could make L-glutamine a potential biomarker for the consumption of these foods. L-Glutamine is a potentially toxic compound. L-Glutamine, with regard to humans, has been found to be associated with several diseases such as schizophrenia, eosinophilic esophagitis, phosphoenolpyruvate carboxykinase deficiency 1, cytosolic, and crohn's disease; L-glutamine has also been linked to the inborn metabolic disorder fumarase deficiency. L-Glutamine is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S)-2,5-Diamino-5-oxopentanoic acid | ChEBI | | (2S)-2-Amino-4-carbamoylbutanoic acid | ChEBI | | (S)-2,5-Diamino-5-oxopentanoic acid | ChEBI | | Glutamic acid 5-amide | ChEBI | | Glutamic acid amide | ChEBI | | GLUTAMINE | ChEBI | | L-(+)-Glutamine | ChEBI | | L-2-Aminoglutaramic acid | ChEBI | | L-Glutamic acid gamma-amide | ChEBI | | L-Glutamin | ChEBI | | L-Glutaminsaeure-5-amid | ChEBI | | Levoglutamide | ChEBI | | Q | ChEBI | | Endari | Kegg | | Nutrestore | Kegg | | (2S)-2,5-Diamino-5-oxopentanoate | Generator | | (2S)-2-Amino-4-carbamoylbutanoate | Generator | | (S)-2,5-Diamino-5-oxopentanoate | Generator | | Glutamate 5-amide | Generator | | Glutamate amide | Generator | | L-2-Aminoglutaramate | Generator | | L-Glutamate g-amide | Generator | | L-Glutamate gamma-amide | Generator | | L-Glutamate γ-amide | Generator | | L-Glutamic acid g-amide | Generator | | L-Glutamic acid γ-amide | Generator | | 2-Aminoglutaramic acid | HMDB | | Cebrogen | HMDB | | gamma-Glutamine | HMDB | | Glavamin | HMDB | | Glumin | HMDB | | L-2-Aminoglutaramidic acid | HMDB | | L-Glutamic acid 5-amide | HMDB | | L-Glutamid | HMDB | | L-Glutamide | HMDB | | Levoglutamid | HMDB | | Levoglutamida | HMDB | | Levoglutamidum | HMDB | | Levoglutamina | HMDB | | Polyglutamine | HMDB | | Stimulina | HMDB | | D-Glutamine | HMDB | | D Glutamine | HMDB | | L Glutamine | HMDB |
|
|---|
| Chemical Formula | C5H10N2O3 |
|---|
| Average Molecular Weight | 146.14 |
|---|
| Monoisotopic Molecular Weight | 146.0691 |
|---|
| IUPAC Name | (2S)-2-amino-4-carbamoylbutanoic acid |
|---|
| Traditional Name | L-glutamine |
|---|
| CAS Registry Number | 56-85-9 |
|---|
| SMILES | N[C@@H](CCC(N)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C5H10N2O3/c6-3(5(9)10)1-2-4(7)8/h3H,1-2,6H2,(H2,7,8)(H,9,10)/t3-/m0/s1 |
|---|
| InChI Key | ZDXPYRJPNDTMRX-VKHMYHEASA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | L-alpha-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - L-alpha-amino acid
- Fatty acid
- Amino acid
- Carboximidic acid
- Carboximidic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Amine
- Hydrocarbon derivative
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Primary aliphatic amine
- Organopnictogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 185 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 41.3 mg/mL | YALKOWSKY,SH & DANNENFELSER,RM (1992) | | logP | -3.64 | CHMELIK,J ET AL. (1991) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-001i-9000000000-bc4e294b8a3fd8c5abce | 2014-09-20 | View Spectrum | | GC-MS | L-Glutamine, 3 TMS, GC-MS Spectrum | splash10-0a4i-0910000000-adb283bb40327f705680 | Spectrum | | GC-MS | L-Glutamine, 3 TMS, GC-MS Spectrum | splash10-0a4i-0910000000-134e80840320dadf6ad1 | Spectrum | | GC-MS | L-Glutamine, 3 TMS, GC-MS Spectrum | splash10-05fr-7910000000-89f87d4acc18299244f5 | Spectrum | | GC-MS | L-Glutamine, 2 TMS, GC-MS Spectrum | splash10-0a4i-0900000000-952a471cad5e5ead0a7e | Spectrum | | GC-MS | L-Glutamine, 4 TMS, GC-MS Spectrum | splash10-004i-1961000000-94183211889cfe72d4ff | Spectrum | | GC-MS | L-Glutamine, 3 TMS, GC-MS Spectrum | splash10-0a4i-1920000000-6505cd814f3707a0feba | Spectrum | | GC-MS | L-Glutamine, 4 TMS, GC-MS Spectrum | splash10-004i-0692000000-9788416fdefb051b9586 | Spectrum | | GC-MS | L-Glutamine, non-derivatized, GC-MS Spectrum | splash10-0a4i-0910000000-adb283bb40327f705680 | Spectrum | | GC-MS | L-Glutamine, non-derivatized, GC-MS Spectrum | splash10-0a4i-0910000000-134e80840320dadf6ad1 | Spectrum | | GC-MS | L-Glutamine, non-derivatized, GC-MS Spectrum | splash10-00dj-4921200000-5446333d9aa592da8a07 | Spectrum | | GC-MS | L-Glutamine, non-derivatized, GC-MS Spectrum | splash10-05fr-7910000000-89f87d4acc18299244f5 | Spectrum | | GC-MS | L-Glutamine, non-derivatized, GC-MS Spectrum | splash10-0a4i-0900000000-952a471cad5e5ead0a7e | Spectrum | | GC-MS | L-Glutamine, non-derivatized, GC-MS Spectrum | splash10-004i-1961000000-94183211889cfe72d4ff | Spectrum | | GC-MS | L-Glutamine, non-derivatized, GC-MS Spectrum | splash10-0a4i-1920000000-6505cd814f3707a0feba | Spectrum | | GC-MS | L-Glutamine, non-derivatized, GC-MS Spectrum | splash10-004i-0692000000-9788416fdefb051b9586 | Spectrum | | GC-MS | L-Glutamine, non-derivatized, GC-MS Spectrum | splash10-0a4i-0910000000-ce2b15fe45c57a30c6bb | Spectrum | | GC-MS | L-Glutamine, non-derivatized, GC-MS Spectrum | splash10-0ab9-1900000000-7be5eab1056d2a249ab5 | Spectrum | | GC-MS | L-Glutamine, non-derivatized, GC-MS Spectrum | splash10-0a4i-0900000000-5dc2d147bea250c7b80e | Spectrum | | GC-MS | L-Glutamine, non-derivatized, GC-MS Spectrum | splash10-0fb9-0921000000-5dcbab01982f543f7925 | Spectrum | | GC-MS | L-Glutamine, non-derivatized, GC-MS Spectrum | splash10-004j-0940000000-102c9a43b5153f06927f | Spectrum | | Predicted GC-MS | L-Glutamine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0006-9300000000-716b1947d48f42862cdf | Spectrum | | Predicted GC-MS | L-Glutamine, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00dl-9610000000-bfd7aa529a6e7f62de43 | Spectrum | | Predicted GC-MS | L-Glutamine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Glutamine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Glutamine, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0059-3900000000-c8f487d1a561e49327c0 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001i-9000000000-eee04836e7577e11d0d0 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0a59-9000000000-54ffbe5be8d7dbede389 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0002-0900000000-79283391c985f8286677 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-9000000000-6afbd5929b6e2371a371 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-d5bd83614703b048228e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-dc8def340a2655f9a399 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0002-0900000000-47e3e94a4387c53e0bd6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-9000000000-c1acfa3750a90d81ce15 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-004i-0900000000-93f01bccbc98f18d4b07 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-ce03708ec9ce55cd8952 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0002-0933201000-9339494cebedd9c39135 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-0900000000-64561a4c3590d5994e37 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0002-0900000000-fdcdd7591fb182d804e2 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-03dm-0030900000-66314ae704783630947a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0002-0933100000-8798d2e46a415bb1029a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-0900000000-dddea137e500a9b364cc | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0002-0900000000-a78f8e4b2d6b37917b21 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0002-0920000000-8c6e1eb7de6f70911080 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0002-0900000000-fa42fc4bcba608992804 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-004i-2900000000-074610880ea19a417f60 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-059x-9200000000-8b7dae54eb8325667bd7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0006-9000000000-e115d9215eea3f0c3629 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0006-9000000000-55b1ac62eea66274b984 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-001j-0900000000-c9c71895033d66cb2bec | 2012-08-31 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 22.5 MHz, D2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY 2D NMR Spectrum (experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Pyrimidine Metabolism |  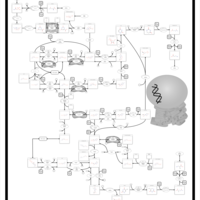  |  | | Glutamate Metabolism | 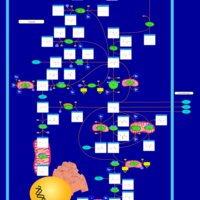 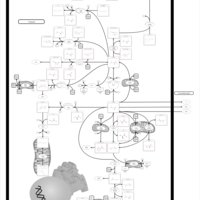 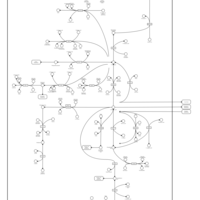 |  | | Transcription/Translation | Not Available | Not Available | | Purine Metabolism |  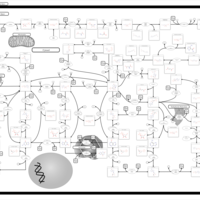 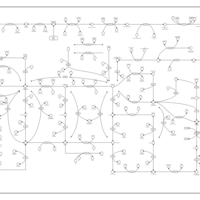 |  | | Urea Cycle | 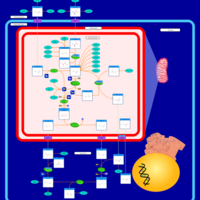 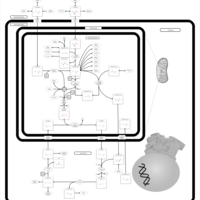 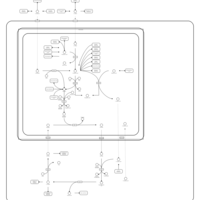 |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 2 | GFPT2 | 5q34-q35 | O94808 | details | | Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | GFPT1 | 2p13 | Q06210 | details | | Amidophosphoribosyltransferase | PPAT | 4q12 | Q06203 | details | | Kynurenine--oxoglutarate transaminase 1 | CCBL1 | 9q34.11 | Q16773 | details | | Glutamine--tRNA ligase | QARS | 3p21.31 | P47897 | details | | Asparagine synthetase [glutamine-hydrolyzing] | ASNS | 7q21.3 | P08243 | details | | GMP synthase [glutamine-hydrolyzing] | GMPS | 3q24 | P49915 | details | | Protein-glutamine gamma-glutamyltransferase E | TGM3 | 20q11.2 | Q08188 | details | | Protein-glutamine gamma-glutamyltransferase 2 | TGM2 | 20q12 | P21980 | details | | Glutamine synthetase | GLUL | 1q31 | P15104 | details | | Glutaminase liver isoform, mitochondrial | GLS2 | 12q13 | Q9UI32 | details | | CTP synthase 1 | CTPS1 | 1p34.1 | P17812 | details | | Protein-glutamine gamma-glutamyltransferase 5 | TGM5 | 15q15.2 | O43548 | details | | Glutaminase kidney isoform, mitochondrial | GLS | 2q32-q34 | O94925 | details | | Protein-glutamine gamma-glutamyltransferase 4 | TGM4 | 3p22-p21.33 | P49221 | details | | Protein-glutamine gamma-glutamyltransferase K | TGM1 | 14q11.2 | P22735 | details | | Protein-glutamine gamma-glutamyltransferase 6 | TGM6 | 20p13 | O95932 | details | | Protein-glutamine gamma-glutamyltransferase Z | TGM7 | 15q15.2 | Q96PF1 | details | | Phosphoribosylformylglycinamidine synthase | PFAS | 17p13.1 | O15067 | details | | Glutamyl-tRNA(Gln) amidotransferase subunit B, mitochondrial | PET112 | 4q31.3 | O75879 | details | | Glutamine-dependent NAD(+) synthetase | NADSYN1 | 11q13.4 | Q6IA69 | details | | CTP synthase 2 | CTPS2 | Xp22 | Q9NRF8 | details | | Glycine N-acyltransferase-like protein 1 | GLYATL1 | 11q12.1 | Q969I3 | details | | Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial | QRSL1 | 6q21 | Q9H0R6 | details | | Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial | GATC | 12q24.31 | O43716 | details |
|
|---|
| Transporters | |
| Neutral amino acid transporter B(0) | SLC1A5 | 19q13.3 | Q15758 | details | | Large neutral amino acids transporter small subunit 2 | SLC7A8 | 14q11.2 | Q9UHI5 | details | | Sodium-coupled neutral amino acid transporter 3 | SLC38A3 | 3p21.3 | Q99624 | details | | Monocarboxylate transporter 10 | SLC16A10 | 6q21-q22 | Q8TF71 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 2.158 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 4.867 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 4.310 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.631 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 2.242 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 5.349 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000641 |
|---|
| DrugBank ID | DB00130 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB030965 |
|---|
| KNApSAcK ID | C00001359 |
|---|
| Chemspider ID | 5746 |
|---|
| KEGG Compound ID | C00064 |
|---|
| BioCyc ID | GLN |
|---|
| BiGG ID | 33714 |
|---|
| Wikipedia Link | Glutamine |
|---|
| METLIN ID | 5614 |
|---|
| PubChem Compound | 5961 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 18050 |
|---|
| References |
|---|
| General References | Not Available |
|---|



