| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:22:45 UTC |
|---|
| Updated at | 2020-12-07 19:06:59 UTC |
|---|
| CannabisDB ID | CDB000045 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Acetaldehyde |
|---|
| Description | Acetaldehyde, also known as ethanal, belongs to the class of organic compounds known as short-chain aldehydes. These are aldehydes with a chain length containing between 2 and 5 carbon atoms. Acetaldehyde is an extremely weak basic (essentially neutral) compound (based on its pKa). Acetaldehyde exists in all living species, ranging from bacteria to humans. Within humans, acetaldehyde participates in a number of enzymatic reactions. In particular, acetaldehyde can be biosynthesized from ethanol; which is mediated by the enzyme alcohol dehydrogenase 1B. In addition, acetaldehyde can be converted into acetic acid; which is mediated by mitochondrial aldehyde dehydrogenases. Industrially, the main method of production of acetaldehyde is via the oxidation of ethylene via the Wacker process. In the 1970s, the world capacity of the Wacker-Hoechst direct oxidation process for producing acetaldehyde exceeded 2 million tonnes annually. Acetaldehyde is found in a number of different foods, such as sweet oranges, pineapples, and mandarin orange (clementine, tangerine), Acetaldehyde has also been detected, but not quantified in, several different foods, such as malabar plums, malus (crabapple), rose hips, natal plums, and medlars. This could make acetaldehyde a potential biomarker for the consumption of these foods. As a food constituent, acetaldehyde is an aldehydic, ethereal, and fruity tasting compound. Acetaldehyde is formally rated as a possible carcinogen (by IARC 2B) and is also a potentially toxic compound. Acetaldehyde has been found to be associated with several diseases such as alcoholism, ulcerative colitis, nonalcoholic fatty liver disease, and Crohn's disease. Acetaldehyde has also been linked to several inborn metabolic disorders including aldehyde dehydrogenase deficiency (III). The level at which an average consumer can detect acetaldehyde is still considerably lower than any toxicity. Pathways of exposure include air, water, land, or groundwater, as well as drink and smoke. Plastics Acetaldehyde is also created by thermal degradation or ultraviolet photo-degradation of some thermoplastic polymers during or after manufacture. Acetaldehyde is known to be the most abundant carcinogen in tobacco smoke; it is easily dissolved in saliva while smoking (PMID:17382522 , PMID:21556207 ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Acetaldehyd | ChEBI | | Acetaldehydes | ChEBI | | Acetic aldehyde | ChEBI | | Azetaldehyd | ChEBI | | Ethanal | ChEBI | | Ethyl aldehyde | ChEBI | | Aldehyde | HMDB |
|
|---|
| Chemical Formula | C2H4O |
|---|
| Average Molecular Weight | 44.05 |
|---|
| Monoisotopic Molecular Weight | 44.0262 |
|---|
| IUPAC Name | acetaldehyde |
|---|
| Traditional Name | acetaldehyde |
|---|
| CAS Registry Number | 75-07-0 |
|---|
| SMILES | CC=O |
|---|
| InChI Identifier | InChI=1S/C2H4O/c1-2-3/h2H,1H3 |
|---|
| InChI Key | IKHGUXGNUITLKF-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as short-chain aldehydes. These are an aldehyde with a chain length containing between 2 and 5 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbonyl compounds |
|---|
| Direct Parent | Short-chain aldehydes |
|---|
| Alternative Parents | |
|---|
| Substituents | - Organic oxide
- Hydrocarbon derivative
- Short-chain aldehyde
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Indirect biological role: Industrial application: Environmental role: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | -123 °C | Not Available | | Boiling Point | 20.2 °C | Wikipedia | | Water Solubility | 1000 mg/mL | Not Available | | logP | -0.34 | TSCATS |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-002f-9000000000-65d53ef91644a0bacd6c | 2014-09-20 | View Spectrum | | Predicted GC-MS | Acetaldehyde, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0006-9000000000-69e31ccd415894a68912 | Spectrum | | Predicted GC-MS | Acetaldehyde, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Acetaldehyde, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0002-9000000000-f1274d4b6066776ca898 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001l-9000000000-c1e37abbf2ad6054dc10 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-000t-9000000000-2289ead4f7210282cd87 | 2012-07-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9000000000-cf54221d95714f5478c4 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9000000000-8d8afe7422ae76f7ebb9 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-004j-9000000000-d68dec9f846cfe9acc72 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9000000000-4430d6a790eca4132aa4 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-607a755de038203a6b68 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-63c9f623d8dc4b1e60a2 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9000000000-00ba25458eb6c0cc2940 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9000000000-00ba25458eb6c0cc2940 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9000000000-0d922cdfd7f6947230c0 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9000000000-452a5f79625d3401d495 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-452a5f79625d3401d495 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-2758497e574a09010547 | 2021-09-23 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 50.18 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Pyruvate Metabolism |  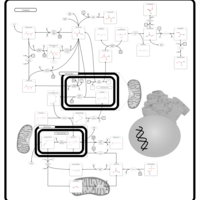  |  | | Ethanol Degradation | 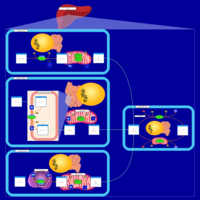 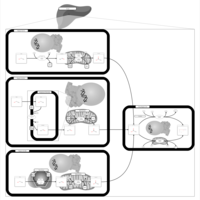 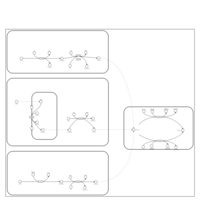 | Not Available | | Disulfiram Action Pathway | 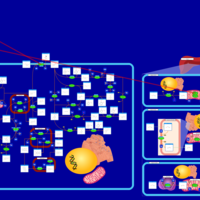 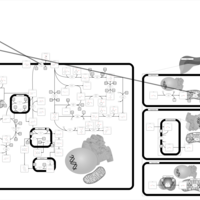 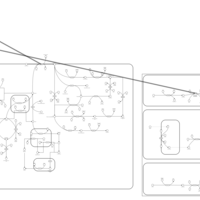 | Not Available | | Pentose Phosphate Pathway | 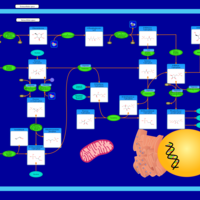  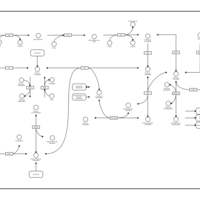 |  | | Glucose-6-phosphate dehydrogenase deficiency | 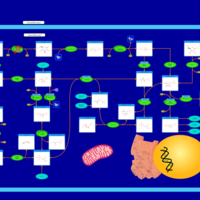 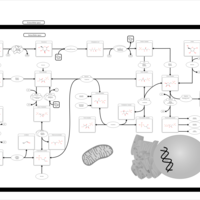 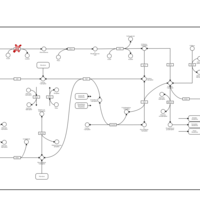 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | |
| Alcohol dehydrogenase class 4 mu/sigma chain | ADH7 | 4q23-q24 | P40394 | details |
|
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000990 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB008297 |
|---|
| KNApSAcK ID | C00007392 |
|---|
| Chemspider ID | 172 |
|---|
| KEGG Compound ID | C00084 |
|---|
| BioCyc ID | ACETALD |
|---|
| BiGG ID | 33792 |
|---|
| Wikipedia Link | Acetaldehyde |
|---|
| METLIN ID | 3200 |
|---|
| PubChem Compound | 177 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15343 |
|---|
| References |
|---|
| General References | - Talhout R, Opperhuizen A, van Amsterdam JG: Role of acetaldehyde in tobacco smoke addiction. Eur Neuropsychopharmacol. 2007 Oct;17(10):627-36. doi: 10.1016/j.euroneuro.2007.02.013. Epub 2007 Mar 23. [PubMed:17382522 ]
- Talhout R, Schulz T, Florek E, van Benthem J, Wester P, Opperhuizen A: Hazardous compounds in tobacco smoke. Int J Environ Res Public Health. 2011 Feb;8(2):613-28. doi: 10.3390/ijerph8020613. Epub 2011 Feb 23. [PubMed:21556207 ]
|
|---|

