| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:44:03 UTC |
|---|
| Updated at | 2020-12-07 19:11:07 UTC |
|---|
| CannabisDB ID | CDB004858 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | L-Methionine |
|---|
| Description | L-Methionine, also known as acimethin or H-met-H, belongs to the class of organic compounds known as methionine and derivatives. Methionine and derivatives are compounds containing methionine or a derivative thereof resulting from reaction of methionine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. The L-enantiomer of methionine. L-Methionine is a drug which is used for protein synthesis including the formation of same, l-homocysteine, l-cysteine, taurine, and sulfate. L-Methionine is a very strong basic compound (based on its pKa). L-Methionine exists in all living species, ranging from bacteria to humans. Outside of the human body, L-Methionine is found, on average, in the highest concentration within a few different foods, such as milk (cow), beluga whales, and smelts and in a lower concentration in white cabbages, wax gourds, and coconuts. L-Methionine has also been detected, but not quantified in, several different foods, such as pepper (c. chinense), acerola, grass pea, oxheart cabbages, and sherries. This could make L-methionine a potential biomarker for the consumption of these foods. L-Methionine is a potentially toxic compound. L-Methionine, with regard to humans, has been found to be associated with several diseases such as autism, leukemia, obesity, and heart failure; L-methionine has also been linked to the inborn metabolic disorder celiac disease. L-Methionine is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S)-2-Amino-4-(methylsulfanyl)butanoic acid | ChEBI | | (S)-2-Amino-4-(methylthio)butanoic acid | ChEBI | | (S)-2-Amino-4-(methylthio)butyric acid | ChEBI | | (S)-Methionine | ChEBI | | L-(-)-Methionine | ChEBI | | L-alpha-Amino-gamma-methylmercaptobutyric acid | ChEBI | | L-Methionin | ChEBI | | M | ChEBI | | Met | ChEBI | | METHIONINE | ChEBI | | L-2-Amino-4methylthiobutyric acid | Kegg | | L-Methionine Z | Kegg | | (2S)-2-Amino-4-(methylsulfanyl)butanoate | Generator | | (2S)-2-Amino-4-(methylsulphanyl)butanoate | Generator | | (2S)-2-Amino-4-(methylsulphanyl)butanoic acid | Generator | | (S)-2-Amino-4-(methylthio)butanoate | Generator | | (S)-2-Amino-4-(methylthio)butyrate | Generator | | L-a-Amino-g-methylmercaptobutyrate | Generator | | L-a-Amino-g-methylmercaptobutyric acid | Generator | | L-alpha-Amino-gamma-methylmercaptobutyrate | Generator | | L-Α-amino-γ-methylmercaptobutyrate | Generator | | L-Α-amino-γ-methylmercaptobutyric acid | Generator | | L-2-Amino-4methylthiobutyrate | Generator | | (L)-Methionine | HMDB | | (S)-(+)-Methionine | HMDB | | (S)-2-Amino-4-(methylthio)-butanoate | HMDB | | (S)-2-Amino-4-(methylthio)-butanoic acid | HMDB | | 2-Amino-4-(methylthio)butyrate | HMDB | | 2-Amino-4-(methylthio)butyric acid | HMDB | | 2-Amino-4-methylthiobutanoate | HMDB | | 2-Amino-4-methylthiobutanoic acid | HMDB | | a-Amino-g-methylmercaptobutyrate | HMDB | | a-Amino-g-methylmercaptobutyric acid | HMDB | | Acimethin | HMDB | | alpha-Amino-alpha-aminobutyric acid | HMDB | | alpha-Amino-gamma-methylmercaptobutyrate | HMDB | | alpha-Amino-gamma-methylmercaptobutyric acid | HMDB | | Cymethion | HMDB | | g-Methylthio-a-aminobutyrate | HMDB | | g-Methylthio-a-aminobutyric acid | HMDB | | gamma-Methylthio-alpha-aminobutyrate | HMDB | | gamma-Methylthio-alpha-aminobutyric acid | HMDB | | H-Met-H | HMDB | | H-Met-OH | HMDB | | L(-)-Amino-alpha-amino-alpha-aminobutyric acid | HMDB | | L(-)-Amino-gamma-methylthiobutyric acid | HMDB | | L-2-Amino-4-(methylthio)butyric acid | HMDB | | L-2-Amino-4-methylthiobutyric acid | HMDB | | L-a-Amino-g-methylthiobutyrate | HMDB | | L-a-Amino-g-methylthiobutyric acid | HMDB | | L-alpha-Amino-gamma-methylthiobutyrate | HMDB | | L-alpha-Amino-gamma-methylthiobutyric acid | HMDB | | L-gamma-Methylthio-alpha-aminobutyric acid | HMDB | | L-Methioninum | HMDB | | Liquimeth | HMDB | | Mepron | HMDB | | Methilanin | HMDB | | Methioninum | HMDB | | Metionina | HMDB | | Neo-methidin | HMDB | | Poly-L-methionine | HMDB | | Polymethionine | HMDB | | S-Methionine | HMDB | | S-Methyl-L-homocysteine | HMDB | | Toxin war | HMDB | | L-Isomer methionine | HMDB | | Methionine, L-isomer | HMDB | | Pedameth | HMDB | | Methionine, L isomer | HMDB | | (3R,3'r,6S)-4,5-DIDEHYDRO-5,6-dihydro-BETA,BETA-carotene-3,3'-diol | ChEBI | | Bo-xan | ChEBI | | e 161b | ChEBI | | Xanthophyll | ChEBI | | (3R,3'r,6S)-4,5-DIDEHYDRO-5,6-dihydro-b,b-carotene-3,3'-diol | Generator | | (3R,3'r,6S)-4,5-DIDEHYDRO-5,6-dihydro-β,β-carotene-3,3'-diol | Generator |
|
|---|
| Chemical Formula | C5H11NO2S |
|---|
| Average Molecular Weight | 149.21 |
|---|
| Monoisotopic Molecular Weight | 149.051 |
|---|
| IUPAC Name | (2S)-2-amino-4-(methylsulfanyl)butanoic acid |
|---|
| Traditional Name | L-methionine |
|---|
| CAS Registry Number | 63-68-3 |
|---|
| SMILES | CSCC[C@H](N)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C5H11NO2S/c1-9-3-2-4(6)5(7)8/h4H,2-3,6H2,1H3,(H,7,8)/t4-/m0/s1 |
|---|
| InChI Key | FFEARJCKVFRZRR-BYPYZUCNSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as methionine and derivatives. Methionine and derivatives are compounds containing methionine or a derivative thereof resulting from reaction of methionine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Methionine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Methionine or derivatives
- Alpha-amino acid
- L-alpha-amino acid
- Thia fatty acid
- Fatty acid
- Fatty acyl
- Amino acid
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Thioether
- Sulfenyl compound
- Dialkylthioether
- Amine
- Organic oxygen compound
- Primary amine
- Organosulfur compound
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Primary aliphatic amine
- Carbonyl group
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Indirect biological role: Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 284 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 56.6 mg/mL | YALKOWSKY,SH & DANNENFELSER,RM (1992) | | logP | -1.87 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | L-Methionine, 2 TMS, GC-MS Spectrum | splash10-004i-0920000000-945c85aa7c9f5eb2dfbb | Spectrum | | GC-MS | L-Methionine, non-derivatized, GC-MS Spectrum | splash10-004i-0910000000-b837ee0f4413856560f1 | Spectrum | | GC-MS | L-Methionine, 2 TMS, GC-MS Spectrum | splash10-00b9-7910000000-5a1558fbb2f5e86edc9b | Spectrum | | GC-MS | L-Methionine, 1 TMS, GC-MS Spectrum | splash10-0udi-1900000000-1a97567ce4f25c4e8263 | Spectrum | | GC-MS | L-Methionine, 2 TMS, GC-MS Spectrum | splash10-004i-0910000000-1b0477118cb20549bf4d | Spectrum | | GC-MS | L-Methionine, non-derivatized, GC-MS Spectrum | splash10-004i-0920000000-8ffae5d87508e0704903 | Spectrum | | GC-MS | L-Methionine, non-derivatized, GC-MS Spectrum | splash10-004i-0920000000-945c85aa7c9f5eb2dfbb | Spectrum | | GC-MS | L-Methionine, non-derivatized, GC-MS Spectrum | splash10-004i-0910000000-b837ee0f4413856560f1 | Spectrum | | GC-MS | L-Methionine, non-derivatized, GC-MS Spectrum | splash10-01bc-2692000000-c6a4af434abaeea2de87 | Spectrum | | GC-MS | L-Methionine, non-derivatized, GC-MS Spectrum | splash10-00b9-7910000000-5a1558fbb2f5e86edc9b | Spectrum | | GC-MS | L-Methionine, non-derivatized, GC-MS Spectrum | splash10-0udi-1900000000-1a97567ce4f25c4e8263 | Spectrum | | GC-MS | L-Methionine, non-derivatized, GC-MS Spectrum | splash10-004i-0910000000-1b0477118cb20549bf4d | Spectrum | | Predicted GC-MS | L-Methionine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0mbd-9200000000-77e5cfb78936ad02d71c | Spectrum | | Predicted GC-MS | L-Methionine, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00di-9510000000-c03dec8575710eed861e | Spectrum | | Predicted GC-MS | L-Methionine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Methionine, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Methionine, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Methionine, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0uea-1900000000-b991859b2c5bed6592a8 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-08fr-9000000000-66855ace60e59837f131 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-08fr-9000000000-56a567791c824c6a9da6 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0fe0-0900000000-d680295f21b2e2b40366 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0006-9000000000-062d3540db4db22da836 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00di-0900000000-84467513e2c9ec1a6851 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-88dc2bebb198eea550ef | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0udi-0920000000-b444ad79abeb16acde43 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-6e24a8df417e5f3db58c | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00di-0900000000-c60ef880eb9a816a274b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-0900000000-a7b7d1a3481c0c691a5b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0002-0900000000-9156f088f4cc9eafa892 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0002-9200000000-f78ba2aab8d5a0e0b135 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0002-9000000000-e7b819fd2d0ac3862860 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0002-9000000000-b61396e720381bd5ff85 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0002-9000000000-b61396e720381bd5ff85 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0udi-0900000000-c3557cb41fd6fe268819 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-0udi-6900000000-6bf5af2d1c561013948a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-08fr-9000000000-63c9b1c138f9a27490e5 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-03di-9000000000-e133c47b0efe4992589f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-03di-9000000000-dee78b4f34f8732fedb0 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - CE-ESI-TOF (CE-system connected to 6210 Time-of-Flight MS, Agilent) , Positive | splash10-0udi-0900000000-d3f03ff5e8eacc8b6c8e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-0udi-0900000000-e0dd5ff44b7962f6a2d2 | 2012-08-31 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-2900000000-305916dde72c899993b6 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-9800000000-47e0a34a2ec02bd762bf | 2016-09-12 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Betaine Metabolism |  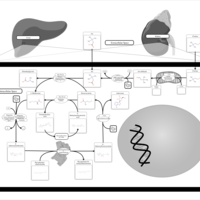  |  | | Methionine Metabolism | 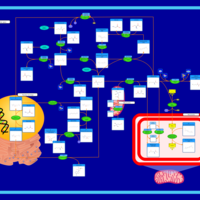 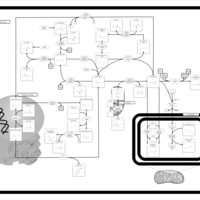 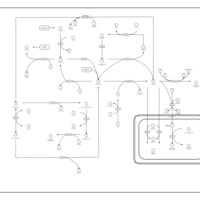 |  | | Transcription/Translation | Not Available | Not Available | | Spermidine and Spermine Biosynthesis | 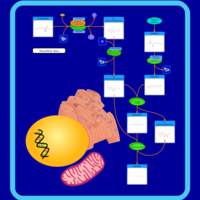 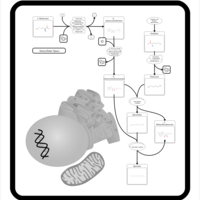 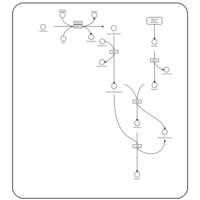 | Not Available | | Glycine and Serine Metabolism | 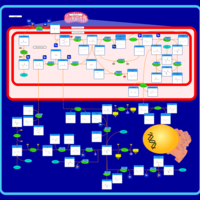 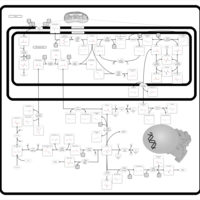 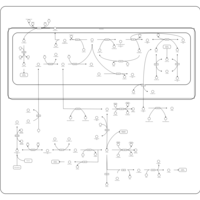 |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | |
| Methionine-R-sulfoxide reductase B2, mitochondrial | MSRB2 | 10p12 | Q9Y3D2 | details |
|
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000696 |
|---|
| DrugBank ID | DB00134 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB012683 |
|---|
| KNApSAcK ID | C00001379 |
|---|
| Chemspider ID | 5907 |
|---|
| KEGG Compound ID | C00073 |
|---|
| BioCyc ID | MET |
|---|
| BiGG ID | 33753 |
|---|
| Wikipedia Link | Methionine |
|---|
| METLIN ID | 5664 |
|---|
| PubChem Compound | 6137 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16643 |
|---|
| References |
|---|
| General References | Not Available |
|---|

