| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:37:57 UTC |
|---|
| Updated at | 2020-12-07 19:10:58 UTC |
|---|
| CannabisDB ID | CDB004799 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Cytidine |
|---|
| Description | Cytidine, also known as Cyd or cytosine riboside, belongs to the class of organic compounds known as pyrimidine nucleosides. Pyrimidine nucleosides are compounds comprising a pyrimidine base attached to a ribosyl or deoxyribosyl moiety. APOBEC is a family of enzymes that has been discovered with the ability to deaminate cytidines on RNA or DNA. Cytidine is a nucleoside that is composed of the base cytosine linked to the five-carbon sugar D-ribose. Cytidine is an extremely weak basic (essentially neutral) compound (based on its pKa). In humans, cytidine is involved in the metabolic disorder called the congenital disorder of glycosylation cdg-iid pathway. An efficient mechanism mediating the brain uptake of circulating cytidine has not yet been demonstrated. The human apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G protein (APOBEC3G, or hA3G), provides cells with an intracellular antiretroviral activity that is associated with the hypermutation of viral DNA through cytidine deamination. It is a precursor of cytidine triphosphate (CTP) needed in the phosphatidylcholine (PC) and phosphatidylethanolamine (PE) biosynthetic pathways. The transport of cytidine into the brain's extracellular fluid, and then into neurons and glia, are essential prerequisites for cytidine to be utilized in the brain. These variations probably reflect the species differences in cytidine deaminase, the enzyme that converts cytidine to uridine in the body. The enzyme that converts CTP to endogenous CDP-choline (CTP:phosphocholine cytidylyltransferase) is unsaturated at physiological brain CTP levels. Cytidine is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-beta-D-Ribofuranosylcytosine | ChEBI | | 1beta-D-Ribofuranosylcytosine | ChEBI | | 4-AMINO-1-BETA-D-ribofuranosyl-2(1H)-pyrimidinone | ChEBI | | 4-Amino-1-beta-D-ribofuranosylpyrimidin-2(1H)-one | ChEBI | | 4-Amino-1beta-D-ribofuranosyl-2(1H)-pyrimidinone | ChEBI | | Cyd | ChEBI | | Cytidin | ChEBI | | Cytosine riboside | ChEBI | | Cytosine-1beta-D-ribofuranoside | ChEBI | | Zytidin | ChEBI | | Posilent | Kegg | | 1-b-D-Ribofuranosylcytosine | Generator | | 1-Β-D-ribofuranosylcytosine | Generator | | 1b-D-Ribofuranosylcytosine | Generator | | 1Β-D-ribofuranosylcytosine | Generator | | 4-AMINO-1-b-D-ribofuranosyl-2(1H)-pyrimidinone | Generator | | 4-AMINO-1-β-D-ribofuranosyl-2(1H)-pyrimidinone | Generator | | 4-Amino-1-b-D-ribofuranosylpyrimidin-2(1H)-one | Generator | | 4-Amino-1-β-D-ribofuranosylpyrimidin-2(1H)-one | Generator | | 4-Amino-1b-D-ribofuranosyl-2(1H)-pyrimidinone | Generator | | 4-Amino-1β-D-ribofuranosyl-2(1H)-pyrimidinone | Generator | | Cytosine-1b-D-ribofuranoside | Generator | | Cytosine-1β-D-ribofuranoside | Generator | | 1-(b-D-Ribofuranosyl)-2-oxo-4-amino-1,2-dihydro-1,3-diazine | HMDB | | 1-(b-delta-Ribofuranosyl)-2-oxo-4-amino-1,2-dihydro-1,3-diazine | HMDB | | 1-b-D-Ribosyl-cytosine | HMDB | | 1-beta-D-Ribofuranosyl-cytosine | HMDB | | 1-beta-delta-Ribofuranosyl-cytosine | HMDB | | 1-beta-delta-Ribofuranosylcytosine | HMDB | | 1-beta-delta-Ribosyl-cytosine | HMDB | | 1beta-delta-Ribofuranosylcytosine | HMDB | | 1beta-Ribofuranosylcytosine | HMDB | | 4-Amino-1-beta-delta-ribofuranosyl-2(1H)-pyrimidinone | HMDB | | Cytosine-1b-delta-ribofuranoside | HMDB | | Cytosine-1beta-delta-ribofuranoside | HMDB | | Cytosine ribonucleoside | HMDB | | Ribonucleoside, cytosine | HMDB | | Riboside, cytosine | HMDB | | 1-(beta-D-Ribofuranosyl)-2-oxo-4-amino-1,2-dihydro-1,3-diazine | HMDB | | 1-(Β-D-ribofuranosyl)-2-oxo-4-amino-1,2-dihydro-1,3-diazine | HMDB |
|
|---|
| Chemical Formula | C9H13N3O5 |
|---|
| Average Molecular Weight | 243.22 |
|---|
| Monoisotopic Molecular Weight | 243.0855 |
|---|
| IUPAC Name | 4-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,2-dihydropyrimidin-2-one |
|---|
| Traditional Name | cytidine |
|---|
| CAS Registry Number | 65-46-3 |
|---|
| SMILES | NC1=NC(=O)N(C=C1)[C@@H]1O[C@H](CO)[C@@H](O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C9H13N3O5/c10-5-1-2-12(9(16)11-5)8-7(15)6(14)4(3-13)17-8/h1-2,4,6-8,13-15H,3H2,(H2,10,11,16)/t4-,6-,7-,8-/m1/s1 |
|---|
| InChI Key | UHDGCWIWMRVCDJ-XVFCMESISA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as pyrimidine nucleosides. Pyrimidine nucleosides are compounds comprising a pyrimidine base attached to a ribosyl or deoxyribosyl moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleosides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Pyrimidine nucleosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine nucleoside
- Glycosyl compound
- N-glycosyl compound
- Pentose monosaccharide
- Aminopyrimidine
- Pyrimidone
- Hydropyrimidine
- Monosaccharide
- Pyrimidine
- Imidolactam
- Tetrahydrofuran
- Heteroaromatic compound
- Secondary alcohol
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Organic nitrogen compound
- Primary alcohol
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Amine
- Alcohol
- Organopnictogen compound
- Organic oxygen compound
- Primary amine
- Hydrocarbon derivative
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 230.5 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | -2.51 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | Cytidine, 5 TMS, GC-MS Spectrum | splash10-0zfr-2951000000-7baf0e1ea8b1a34362e2 | Spectrum | | GC-MS | Cytidine, 4 TMS, GC-MS Spectrum | splash10-0g4j-2980000000-9337623aef7fec22dd97 | Spectrum | | GC-MS | Cytidine, non-derivatized, GC-MS Spectrum | splash10-0zfr-2951000000-7baf0e1ea8b1a34362e2 | Spectrum | | GC-MS | Cytidine, non-derivatized, GC-MS Spectrum | splash10-0g4j-2980000000-9337623aef7fec22dd97 | Spectrum | | Predicted GC-MS | Cytidine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-05dl-9420000000-f2b3f5707444409e125d | Spectrum | | Predicted GC-MS | Cytidine, 3 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0083-6964300000-64e7e138ddd30a0aec2c | Spectrum | | Predicted GC-MS | Cytidine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_1_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_2_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_2_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_2_6, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_2_7, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_3_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_3_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_3_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_3_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_3_6, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TMS_3_7, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cytidine, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-03di-0900000000-177050786fcbfaf2d270 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-03di-0900000000-1251248af66dca3401e2 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-03di-6900000000-c6b010a1d3e65a0942ee | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0006-0590100000-94eb2baae24aa405b1ba | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0006-9000000000-8155621584c25975ffa1 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-03di-0900000000-5065bc83a32bee37d07c | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0006-0590200000-4d5dc5376c40d083efbe | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-6ea86f62a48910b456a0 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-03di-0900000000-5c93b66b8899a4d3ce06 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0006-0090000000-f0e6ac0bacdbef1206b0 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-000f-0090010000-1cf915fa4dfb86b5861b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0a4i-0900000000-6bb7990f165932decea7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0006-0090000000-0b7dc368d61ee31afb78 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-000l-0090020000-fa04fb11f98259c403b6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0006-0190172001-0671df3c315b8af9e13f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0a4i-0900000000-8aa3fec051a7d9807888 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0006-0090000000-0563823e8f3a9def519a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-0028900000-f4b6c319a7e9fb17df2e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-01ox-0590000000-d5d20f84ab30e96c30f6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) 30V, Positive | splash10-03di-0930000000-c4d3377790616b8b7869 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0a4i-1930000000-faec28788ea577988b82 | 2012-08-31 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0910000000-9575881412c4c94a7eae | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-5900000000-2cb5f7e3dbd7baa35b56 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dj-9600000000-7d281b2b2d57c8786e11 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01ox-0960000000-db9207082a1fabf42dae | 2016-09-12 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Pyrimidine Metabolism |  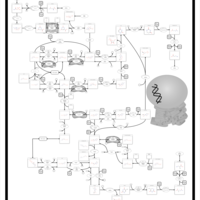  |  | | Beta Ureidopropionase Deficiency |  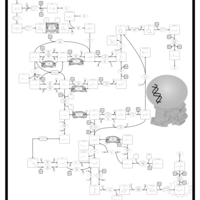 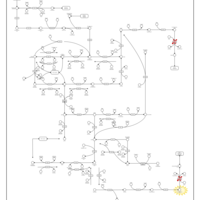 | Not Available | | UMP Synthase Deficiency (Orotic Aciduria) |  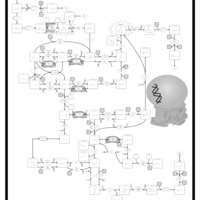  | Not Available | | Dihydropyrimidinase Deficiency |    | Not Available | | MNGIE (Mitochondrial Neurogastrointestinal Encephalopathy) |   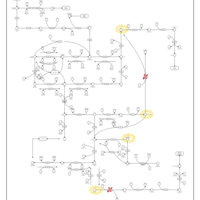 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000089 |
|---|
| DrugBank ID | DB02097 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB021809 |
|---|
| KNApSAcK ID | C00042440 |
|---|
| Chemspider ID | 5940 |
|---|
| KEGG Compound ID | C00475 |
|---|
| BioCyc ID | CYTIDINE |
|---|
| BiGG ID | 35089 |
|---|
| Wikipedia Link | Cytidine |
|---|
| METLIN ID | 3376 |
|---|
| PubChem Compound | 6175 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17562 |
|---|
| References |
|---|
| General References | Not Available |
|---|
