| Identification |
|---|
| HMDB Protein ID
| CDBP02455 |
| Secondary Accession Numbers
| Not Available |
| Name
| Cytidine deaminase |
| Description
| Not Available |
| Synonyms
|
- Cytidine aminohydrolase
|
| Gene Name
| CDA |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in zinc ion binding |
| Specific Function
| This enzyme scavenge exogenous and endogenous cytidine and 2'-deoxycytidine for UMP synthesis.
|
| GO Classification
|
| Biological Process |
| cell surface receptor signaling pathway |
| negative regulation of cell growth |
| protein homotetramerization |
| cellular response to external biotic stimulus |
| cytosine metabolic process |
| negative regulation of nucleotide metabolic process |
| pyrimidine nucleoside salvage |
| response to cycloheximide |
| Cellular Component |
| cytosol |
| extracellular region |
| Function |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| catalytic activity |
| hydrolase activity |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds |
| transition metal ion binding |
| zinc ion binding |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines |
| cytidine deaminase activity |
| Molecular Function |
| nucleoside binding |
| metal ion binding |
| cytidine deaminase activity |
| zinc ion binding |
| protein homodimerization activity |
| Process |
| metabolic process |
| nitrogen compound metabolic process |
| cellular nitrogen compound metabolic process |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| nucleobase, nucleoside and nucleotide metabolic process |
| nucleoside metabolic process |
| pyrimidine nucleoside metabolic process |
| pyrimidine ribonucleoside metabolic process |
| cytidine metabolic process |
|
| Cellular Location
|
- Cytoplasmic
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Drug metabolism - other enzymes | Not Available | 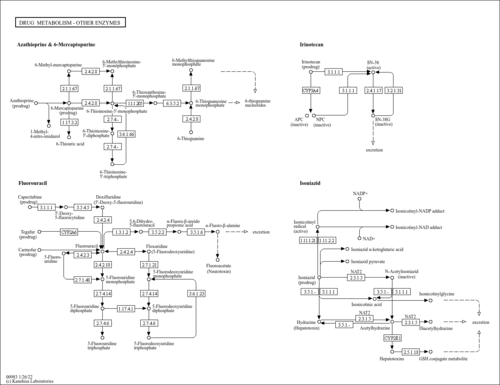 | | Pyrimidine Metabolism |    |  | | Beta Ureidopropionase Deficiency |    | Not Available | | UMP Synthase Deficiency (Orotic Aciduria) |  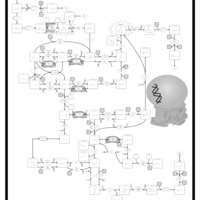 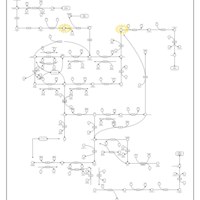 | Not Available | | Dihydropyrimidinase Deficiency |  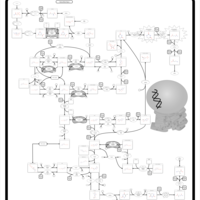 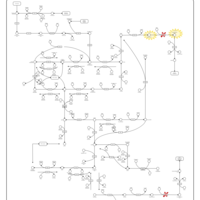 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 1 |
| Locus
| 1p36.2-p35 |
| SNPs
| CDA |
| Gene Sequence
|
>441 bp
ATGGCCCAGAAGCGTCCTGCCTGCACCCTGAAGCCTGAGTGTGTCCAGCAGCTGCTGGTT
TGCTCCCAGGAGGCCAAGAAGTCAGCCTACTGCCCCTACAGTCACTTTCCTGTGGGGGCT
GCCCTGCTCACCCAGGAGGGGAGAATCTTCAAAGGGTGCAACATAGAAAATGCCTGCTAC
CCGCTGGGCATCTGTGCTGAACGGACCGCTATCCAGAAGGCCGTCTCAGAAGGGTACAAG
GATTTCAGGGCAATTGCTATCGCCAGTGACATGCAAGATGATTTTATCTCTCCATGTGGG
GCCTGCAGGCAAGTCATGAGAGAGTTTGGCACCAACTGGCCCGTGTACATGACCAAGCCG
GATGGTACGTATATTGTCATGACGGTCCAGGAGCTGCTGCCCTCCTCCTTTGGGCCTGAG
GACCTGCAGAAGACTCAGTGA
|
| Protein Properties |
|---|
| Number of Residues
| 146 |
| Molecular Weight
| 16184.545 |
| Theoretical pI
| 6.921 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Cytidine deaminase
MAQKRPACTLKPECVQQLLVCSQEAKKSAYCPYSHFPVGAALLTQEGRIFKGCNIENACY
PLGICAERTAIQKAVSEGYKDFRAIAIASDMQDDFISPCGACRQVMREFGTNWPVYMTKP
DGTYIVMTVQELLPSSFGPEDLQKTQ
|
| External Links |
|---|
| GenBank ID Protein
| 598149 |
| UniProtKB/Swiss-Prot ID
| P32320 |
| UniProtKB/Swiss-Prot Entry Name
| CDD_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| L27943 |
| GeneCard ID
| CDA |
| GenAtlas ID
| CDA |
| HGNC ID
| HGNC:1712 |
| References |
|---|
| General References
| Not Available |

