| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:36:56 UTC |
|---|
| Updated at | 2020-12-07 19:10:56 UTC |
|---|
| CannabisDB ID | CDB004789 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Ascorbic acid |
|---|
| Description | Ascorbic acid, also known as acide ascorbique or L-ascorbate, belongs to the class of organic compounds known as butenolides. These are dihydrofurans with a carbonyl group at the C2 carbon atom. Ascorbic acid is a drug which is used to treat vitamin c deficiency, scurvy, delayed wound and bone healing, urine acidification, and in general as an antioxidant. it has also been suggested to be an effective antiviral agent. Ascorbic acid is an extremely weak basic (essentially neutral) compound (based on its pKa). dopamine and ascorbic acid can be converted into norepinephrine and dehydroascorbic acid through the action of the enzyme dopamine beta-hydroxylase. In humans, ascorbic acid is involved in the metabolic disorder called tyrosinemia, transient, of the newborn. Ascorbic acid is a very mild and grassy tasting compound. Outside of the human body, Ascorbic acid is found, on average, in the highest concentration within a few different foods, such as acerola, pepper (c. frutescens), and orange bell peppers and in a lower concentration in yogurts, yardlong beans, and peanuts. Ascorbic acid has also been detected, but not quantified in, several different foods, such as deerberries, cetacea (dolphin, porpoise, whale), acorns, lemon balms, and gelatins. This could make ascorbic acid a potential biomarker for the consumption of these foods. The L-enantiomer of ascorbic acid and conjugate acid of L-ascorbate. Ascorbic acid is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Acide ascorbique | ChEBI | | Acido ascorbico | ChEBI | | Acidum ascorbicum | ChEBI | | Acidum ascorbinicum | ChEBI | | Ascoltin | ChEBI | | Ascorbicap | ChEBI | | Ascorbinsaeure | ChEBI | | e 300 | ChEBI | | e-300 | ChEBI | | e300 | ChEBI | | L-(+)-Ascorbic acid | ChEBI | | L-Ascorbate | ChEBI | | Vitamin C | ChEBI | | Monodehydroascorbate radical | Kegg | | Ascorbate radical | Kegg | | Semidehydroascorbic acid | Kegg | | ASCOR | Kegg | | L-(+)-Ascorbate | Generator | | L-Ascorbic acid | Generator | | Monodehydroascorbic acid radical | Generator | | Ascorbic acid radical | Generator | | Semidehydroascorbate | Generator | | Ascorbate | Generator | | Ascorbic acid, monosodium salt | MeSH | | Ferrous ascorbate | MeSH | | L Ascorbic acid | MeSH | | Magnesium di L ascorbate | MeSH | | Magnorbin | MeSH | | Sodium ascorbate | MeSH | | Acid, L-ascorbic | MeSH | | Ascorbate, magnesium | MeSH | | Acid, ascorbic | MeSH | | Ascorbate, ferrous | MeSH | | Ascorbate, sodium | MeSH | | Hybrin | MeSH | | Magnesium ascorbate | MeSH | | Magnesium ascorbicum | MeSH | | Magnesium di-L-ascorbate | MeSH | | Di-L-ascorbate, magnesium | MeSH | | (+)-Ascorbate | HMDB | | (+)-Ascorbic acid | HMDB | | (+)-Sodium L-ascorbate | HMDB | | 3-keto-L-Gulofuranolactone | HMDB | | 3-oxo-L-Gulofuranolactone | HMDB | | Adenex | HMDB | | Allercorb | HMDB | | Antiscorbic vitamin | HMDB | | Antiscorbutic vitamin | HMDB | | arco-Cee | HMDB | | Ascor-b.i.d. | HMDB | | Ascorb | HMDB | | Ascorbajen | HMDB | | Ascorbicab | HMDB | | Ascorbicin | HMDB | | Ascorbin | HMDB | | Ascorbutina | HMDB | | Ascorin | HMDB | | Ascorteal | HMDB | | Ascorvit | HMDB | | C-Level | HMDB | | C-Long | HMDB | | C-Quin | HMDB | | C-Span | HMDB | | C-Vimin | HMDB | | Cantan | HMDB | | Cantaxin | HMDB | | Catavin C | HMDB | | Ce lent | HMDB | | Ce-mi-lin | HMDB | | Ce-vi-sol | HMDB | | Cebicure | HMDB | | Cebid | HMDB | | Cebion | HMDB | | Cebione | HMDB | | Cecon | HMDB | | Cee-caps TD | HMDB | | Cee-vite | HMDB | | Cegiolan | HMDB | | Ceglion | HMDB | | Ceklin | HMDB | | Celaskon | HMDB | | Celin | HMDB | | Cell C | HMDB | | Cemagyl | HMDB | | Cemill | HMDB | | Cenetone | HMDB | | Cenolate | HMDB | | Cereon | HMDB | | Cergona | HMDB | | Cescorbat | HMDB | | Cetamid | HMDB | | Cetane | HMDB | | Cetane-caps TC | HMDB | | Cetane-caps TD | HMDB | | Cetebe | HMDB | | Cetemican | HMDB | | Cevalin | HMDB | | Cevatine | HMDB | | Cevex | HMDB | | Cevi-bid | HMDB | | Cevimin | HMDB | | Cevital | HMDB | | Cevitamate | HMDB | | Cevitamic acid | HMDB | | Cevitamin | HMDB | | Cevitan | HMDB | | Cevitex | HMDB | | Cewin | HMDB | | Chewcee | HMDB | | Ciamin | HMDB | | Cipca | HMDB | | Citriscorb | HMDB | | Citrovit | HMDB | | Colascor | HMDB | | Concemin | HMDB | | Davitamon C | HMDB | | Dora-C-500 | HMDB | | Duoscorb | HMDB | | gamma-Lactone L-threo-hex-2-enonate | HMDB | | gamma-Lactone L-threo-hex-2-enonic acid | HMDB | | HiCee | HMDB | | ido-C | HMDB | | Juvamine | HMDB | | Kangbingfeng | HMDB | | Kyselina askorbova | HMDB | | L(+)-Ascorbate | HMDB | | L(+)-Ascorbic acid | HMDB | | L-3-Ketothreohexuronic acid lactone | HMDB | | L-Lyxoascorbate | HMDB | | L-Lyxoascorbic acid | HMDB | | L-threo-Ascorbic acid | HMDB | | L-Xyloascorbate | HMDB | | L-Xyloascorbic acid | HMDB | | Laroscorbine | HMDB | | Lemascorb | HMDB | | Liqui-cee | HMDB | | Meri-c | HMDB | | Natrascorb | HMDB | | Natrascorb injectable | HMDB | | Planavit C | HMDB | | Proscorbin | HMDB | | Redoxon | HMDB | | Ribena | HMDB | | Ronotec 100 | HMDB | | Rontex 100 | HMDB | | Roscorbic | HMDB | | Rovimix C | HMDB | | Scorbacid | HMDB | | Scorbu C | HMDB | | Scorbu-C | HMDB | | Secorbate | HMDB | | Sodascorbate | HMDB | | Suncoat VC 40 | HMDB | | Testascorbic | HMDB | | VASC | HMDB | | Vicelat | HMDB | | Vicin | HMDB | | Vicomin C | HMDB | | Viforcit | HMDB | | Viscorin | HMDB | | Viscorin 100m | HMDB | | Vitace | HMDB | | Vitacee | HMDB | | Vitacimin | HMDB | | Vitacin | HMDB | | Vitamisin | HMDB | | Vitascorbol | HMDB | | Xitix | HMDB | | ASCORBIC ACID | ChEBI |
|
|---|
| Chemical Formula | C6H8O6 |
|---|
| Average Molecular Weight | 176.12 |
|---|
| Monoisotopic Molecular Weight | 176.0321 |
|---|
| IUPAC Name | (5R)-5-[(1S)-1,2-dihydroxyethyl]-3,4-dihydroxy-2,5-dihydrofuran-2-one |
|---|
| Traditional Name | vitamin C |
|---|
| CAS Registry Number | 50-81-7 |
|---|
| SMILES | [H][C@@]1(OC(=O)C(O)=C1O)[C@@H](O)CO |
|---|
| InChI Identifier | InChI=1S/C6H8O6/c7-1-2(8)5-3(9)4(10)6(11)12-5/h2,5,7-10H,1H2/t2-,5+/m0/s1 |
|---|
| InChI Key | CIWBSHSKHKDKBQ-JLAZNSOCSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as butenolides. These are dihydrofurans with a carbonyl group at the C2 carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Dihydrofurans |
|---|
| Sub Class | Furanones |
|---|
| Direct Parent | Butenolides |
|---|
| Alternative Parents | |
|---|
| Substituents | - 2-furanone
- Vinylogous acid
- Alpha,beta-unsaturated carboxylic ester
- Enoate ester
- 1,2-diol
- Carboxylic acid ester
- Enediol
- Secondary alcohol
- Lactone
- Carboxylic acid derivative
- Oxacycle
- Monocarboxylic acid or derivatives
- Alcohol
- Hydrocarbon derivative
- Organic oxygen compound
- Carbonyl group
- Organic oxide
- Primary alcohol
- Organooxygen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Biological role: Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 191 °C | PhysProp | | Boiling Point | Not Available | Not Available | | Water Solubility | 400 mg/mL at 40 °C | MERCK INDEX (1996) | | logP | -1.85 | AVDEEF,A (1997) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | Ascorbic acid, non-derivatized, GC-MS Spectrum | splash10-004i-4900000000-e99089fd55560fb70cfe | Spectrum | | GC-MS | Ascorbic acid, non-derivatized, GC-MS Spectrum | splash10-0002-0912000000-baa8cffda0478e1bc197 | Spectrum | | GC-MS | Ascorbic acid, non-derivatized, GC-MS Spectrum | splash10-00di-9400000000-8ff8f48bfa2dd4fd3cf5 | Spectrum | | GC-MS | Ascorbic acid, non-derivatized, GC-MS Spectrum | splash10-00di-9400000000-c1dc72c6cbc49fbf3454 | Spectrum | | GC-MS | Ascorbic acid, non-derivatized, GC-MS Spectrum | splash10-00di-9400000000-b207f4024993c5a74769 | Spectrum | | GC-MS | Ascorbic acid, non-derivatized, GC-MS Spectrum | splash10-0002-0911000000-9af0c08e85e0c51a25c4 | Spectrum | | GC-MS | Ascorbic acid, non-derivatized, GC-MS Spectrum | splash10-004i-4900000000-e99089fd55560fb70cfe | Spectrum | | GC-MS | Ascorbic acid, non-derivatized, GC-MS Spectrum | splash10-0002-0912000000-baa8cffda0478e1bc197 | Spectrum | | GC-MS | Ascorbic acid, non-derivatized, GC-MS Spectrum | splash10-00di-9400000000-8ff8f48bfa2dd4fd3cf5 | Spectrum | | GC-MS | Ascorbic acid, non-derivatized, GC-MS Spectrum | splash10-00di-9400000000-c1dc72c6cbc49fbf3454 | Spectrum | | GC-MS | Ascorbic acid, non-derivatized, GC-MS Spectrum | splash10-00di-9400000000-b207f4024993c5a74769 | Spectrum | | GC-MS | Ascorbic acid, non-derivatized, GC-MS Spectrum | splash10-0002-0911000000-9af0c08e85e0c51a25c4 | Spectrum | | Predicted GC-MS | Ascorbic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0560-9700000000-ab53eee999ee7fee672d | Spectrum | | Predicted GC-MS | Ascorbic acid, 4 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0kkj-9357500000-5c2c439f726b729dd50d | Spectrum | | Predicted GC-MS | Ascorbic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Ascorbic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Ascorbic acid, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Ascorbic acid, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Ascorbic acid, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Ascorbic acid, TMS_1_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Ascorbic acid, TMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Ascorbic acid, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Ascorbic acid, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Ascorbic acid, TMS_2_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Ascorbic acid, TMS_2_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - NA , positive | splash10-0002-9300000000-d8c8254ad1dd4de2674c | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 35V, positive | splash10-004i-1900000000-da5bc92501d37735007f | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - NA , positive | splash10-0006-0900000000-b653774b8718c29fe97a | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-000i-9200000000-5d1ea04ef6404f5ecf7c | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 35V, negative | splash10-000i-9000000000-0371d7191fa8a89176f3 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 6V, negative | splash10-004i-0900000000-76825c62319d5f4cbed9 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 10V, negative | splash10-016r-2900000000-700d5d6247efb36c48d7 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 13V, negative | splash10-014i-8900000000-4d76fc951d56a75a025e | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 16V, negative | splash10-05tr-9300000000-0967cf71e141f71c2e00 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QqQ 20V, negative | splash10-052r-9100000000-2bca6a943255612f1e7d | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 12V, negative | splash10-000i-9000000000-804b40cc657a43d298fb | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 12V, negative | splash10-0a4i-9000000000-cad0cc415c906a783cfc | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 12V, negative | splash10-0a4r-9000000000-f79a7dfc866621b3c926 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 2V, negative | splash10-004i-0900000000-707796161cd4fae98f67 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 2V, negative | splash10-004i-0900000000-035a78987127cc9f23f5 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 2V, negative | splash10-004i-0900000000-dac785ef05051971e946 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 3V, negative | splash10-00or-1900000000-a91dc1a29cb724a17811 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 4V, negative | splash10-016r-3900000000-78f1f46067d2459f17c7 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 5V, negative | splash10-014i-6900000000-8044f242bb90f54a595a | 2020-07-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0kdi-2900000000-f05a720e6e6361c97e5f | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0bvl-5900000000-6ca3f37a74403d8495c9 | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9100000000-5625440c512f60760fdf | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0o90-2900000000-b7c897849334927f5bf5 | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0bvi-4900000000-1f1fa10e7e06a79c028a | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-08fu-9300000000-b98375c0cd3a54b61ef7 | 2016-08-03 | View Spectrum |
|
|---|
| NMR | Not Available |
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Catecholamine Biosynthesis | 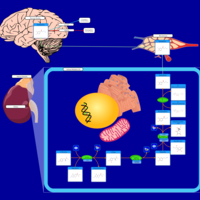 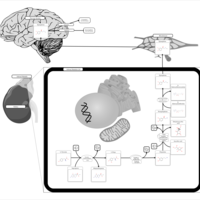  |  | | Carnitine Synthesis | 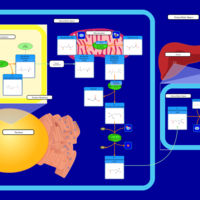 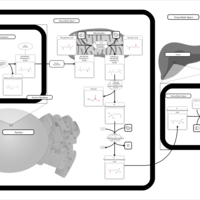 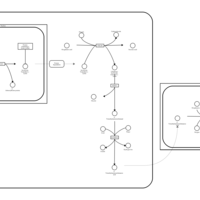 | Not Available | | Aromatic L-Aminoacid Decarboxylase Deficiency | 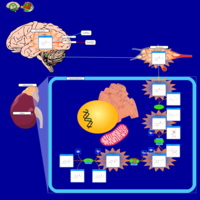 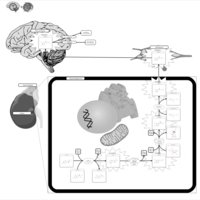  | Not Available | | Tyrosine hydroxylase deficiency |    | Not Available | | Phytanic Acid Peroxisomal Oxidation |   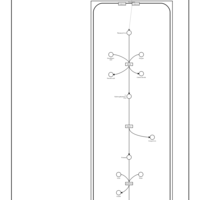 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0014273 |
|---|
| DrugBank ID | DB00126 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB001224 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| KEGG Compound ID | C01041 |
|---|
| BioCyc ID | ASCORBATE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Ascorbic_Acid |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 54670067 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 29073 |
|---|
| References |
|---|
| General References | Not Available |
|---|
