| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-30 17:44:01 UTC |
|---|
| Updated at | 2020-12-07 19:07:47 UTC |
|---|
| CannabisDB ID | CDB000806 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | PA(16:0/16:0) |
|---|
| Description | PA(16:0/16:0)is a phosphatidic acid. It is a glycerophospholipid in which a phosphate moiety occupies a glycerol substitution site. As is the case with diacylglycerols, phosphatidic acids can have many different combinations of fatty acids of varying lengths and saturation attached at the C-1 and C-2 positions. Fatty acids containing 16, 18 and 20 carbons are the most common. PA(16:0/16:0), in particular, consists of two hexadecanoyl chain at positions C-1 and C2. The oleic acid moiety is derived from vegetable oils, especially olive and canola oil, while the oleic acid moiety is derived from vegetable oils, especially olive and canola oil. Phosphatidic acids are quite rare but are extremely important as intermediates in the biosynthesis of triacylglycerols and phospholipids. This compound is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,2-Dipalmitoyl-3-sn-phosphatidic acid | ChEBI | | 1,2-Dipalmitoyl-sn-glycerol-3-phosphate | ChEBI | | 1,2-Dipalmitoyl-sn-glycerol-3-phosphoric acid | ChEBI | | Dipalmitoyl phosphatidic acid | ChEBI | | PA(32:0) | ChEBI | | Phosphatidic acid(16:0/16:0) | ChEBI | | Phosphatidic acid(32:0) | ChEBI | | 1,2-Dipalmitoyl-3-sn-phosphatidate | Generator | | Dipalmitoyl phosphatidate | Generator | | Phosphatidate(16:0/16:0) | Generator | | Phosphatidate(32:0) | Generator | | 1,2-Dipalmitoyl-sn-glycero-3-phosphate | MeSH | | Dipalmitoylphosphatidic acid | MeSH | | Dipalmitoylphosphatidic acid, (+-)-isomer | MeSH | | Dipalmitoylphosphatidic acid, (R)-isomer | MeSH | | Dipalmitoylphosphatidic acid, ammonium salt | MeSH | | Dipalmitoylphosphatidic acid, calcium salt | MeSH | | Dipalmitoylphosphatidic acid, sodium salt | MeSH | | 1,2-Di-O-palmitoyl-3-sn-glyceryl-O-phosphorate | HMDB | | 1,2-Di-O-palmitoyl-3-sn-glyceryl-O-phosphoric acid | HMDB | | 1,2-Dihexadecanoyl-rac-phosphatidic acid | HMDB | | 1,2-Dipalmitoyl-sn-glycerol 3-phosphate | HMDB | | 1,2-Dipalmitoyl-sn-glycerol-3-phosphorate | HMDB | | Dipalmitoyl-L-a-phosphatidate | HMDB | | Dipalmitoyl-L-a-phosphatidic acid | HMDB | | Dipalmitoyl-L-alpha-phosphatidate | HMDB | | Dipalmitoyl-L-alpha-phosphatidic acid | HMDB | | Dipalmitoylphosphatidate | HMDB | | L-a-Dipalmitoyl-phosphatidate | HMDB | | L-a-Dipalmitoyl-phosphatidic acid | HMDB | | L-a-Dipalmitoylphosphatidate | HMDB | | L-a-Dipalmitoylphosphatidic acid | HMDB | | L-alpha-Dipalmitoyl-phosphatidate | HMDB | | L-alpha-Dipalmitoyl-phosphatidic acid | HMDB | | L-alpha-Dipalmitoylphosphatidate | HMDB | | L-alpha-Dipalmitoylphosphatidic acid | HMDB |
|
|---|
| Chemical Formula | C35H69O8P |
|---|
| Average Molecular Weight | 648.9 |
|---|
| Monoisotopic Molecular Weight | 648.473 |
|---|
| IUPAC Name | [(2R)-2,3-bis(hexadecanoyloxy)propoxy]phosphonic acid |
|---|
| Traditional Name | dipalmitoyl |
|---|
| CAS Registry Number | 7091-44-3 |
|---|
| SMILES | [H][C@@](COC(=O)CCCCCCCCCCCCCCC)(COP(O)(O)=O)OC(=O)CCCCCCCCCCCCCCC |
|---|
| InChI Identifier | InChI=1S/C35H69O8P/c1-3-5-7-9-11-13-15-17-19-21-23-25-27-29-34(36)41-31-33(32-42-44(38,39)40)43-35(37)30-28-26-24-22-20-18-16-14-12-10-8-6-4-2/h33H,3-32H2,1-2H3,(H2,38,39,40)/t33-/m1/s1 |
|---|
| InChI Key | PORPENFLTBBHSG-MGBGTMOVSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as 1,2-diacylglycerol-3-phosphates. These are glycerol-3-phosphates in which the glycerol moiety is bonded to two aliphatic chains through ester linkages. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphates |
|---|
| Direct Parent | 1,2-diacylglycerol-3-phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1,2-diacylglycerol-3-phosphate
- Fatty acid ester
- Monoalkyl phosphate
- Dicarboxylic acid or derivatives
- Fatty acyl
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Carboxylic acid ester
- Carboxylic acid derivative
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Organooxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Organoleptic effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 161 - 165 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | PA(16:0/16:0), non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0005-9236341000-d0296e5b3dd53599a39d | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-0002-0000009000-b3b708036c56c15fec89 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-0a4i-1492100000-5624ed03f664b67f79ec | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-0a4i-1490000000-1ba8a217ef22599ee07c | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0002-0002009000-7bdcaa7752c2e2970fdf | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0000900000-42187062de8aa3a7fcef | 2016-09-19 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a71-4092103000-08a70ad05021be172e3b | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9050000000-dc7f569f98cd450257d0 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-f628227e9a0ceffbeaf0 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0000009000-8e723670c2d6da7d1500 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-0000099000-cef7e1d91751b702106e | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00xr-0000946000-9592d132939363ae4ad4 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0000009000-5d6932d42201ac99e40a | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-052e-1166109000-3a40d8a06827026e1331 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-1191101000-55ca7d5b11a7d0030aea | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001j-0000009000-6c5d51790d44b8f28697 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0f6t-0000059000-f0c15a320524af660ac4 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udl-0006093000-7e3d155745f467b43ae7 | 2021-09-25 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, CD3OD, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Phospholipid Biosynthesis |  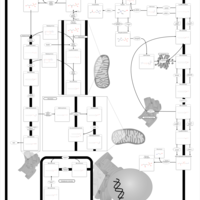 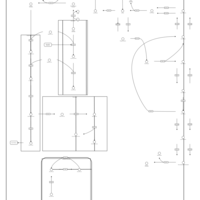 |  | | Glycerolipid Metabolism | 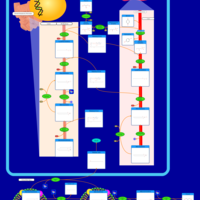 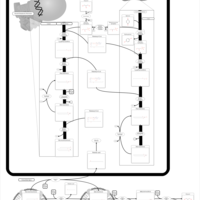 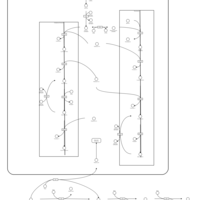 |  | | Glycerol Kinase Deficiency | 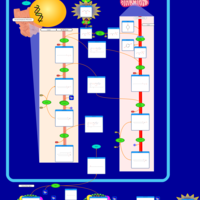 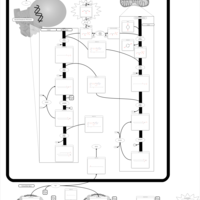 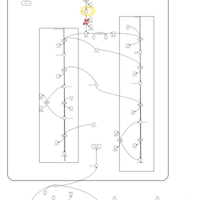 | Not Available | | D-glyceric acidura | 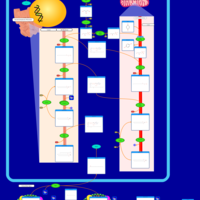 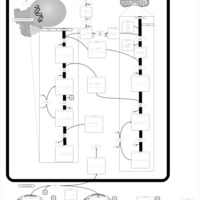 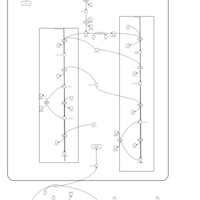 | Not Available | | Familial lipoprotein lipase deficiency | 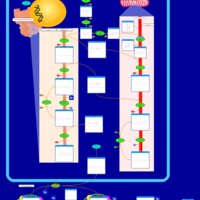   | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Phospholipase D2 | PLD2 | 17p13.1 | O14939 | details | | Diacylglycerol kinase theta | DGKQ | 4p16.3 | P52824 | details | | Diacylglycerol kinase gamma | DGKG | 3q27.2-q27.3 | P49619 | details | | Lipid phosphate phosphohydrolase 2 | PPAP2C | 19p13 | O43688 | details | | Diacylglycerol kinase alpha | DGKA | 12q13.3 | P23743 | details | | Diacylglycerol kinase delta | DGKD | 2q37.1 | Q16760 | details | | Lipid phosphate phosphohydrolase 1 | PPAP2A | 5q11 | O14494 | details | | Diacylglycerol kinase beta | DGKB | 7p21.2 | Q9Y6T7 | details | | Diacylglycerol kinase iota | DGKI | 7q32.3-q33 | O75912 | details | | Lipid phosphate phosphohydrolase 3 | PPAP2B | 1p32.2 | O14495 | details | | Diacylglycerol kinase zeta | DGKZ | 11p11.2 | Q13574 | details | | Phospholipase DDHD1 | DDHD1 | 14q21 | Q8NEL9 | details | | cAMP-specific 3',5'-cyclic phosphodiesterase 4D | PDE4D | 5q12 | Q08499 | details | | Phosphatidate cytidylyltransferase 2 | CDS2 | 20p13 | O95674 | details | | Phosphatidate cytidylyltransferase 1 | CDS1 | 4q21.23 | Q92903 | details | | Probable phospholipid-transporting ATPase IG | ATP11C | | Q8NB49 | details | | Probable phospholipid-transporting ATPase IH | ATP11A | 13q34 | P98196 | details | | Probable phospholipid-transporting ATPase VA | ATP10A | 15q11.2 | O60312 | details | | Probable phospholipid-transporting ATPase IC | ATP8B1 | 18q21-q22|18q21.31 | O43520 | details | | Probable phospholipid-transporting ATPase IIA | ATP9A | 20q13.2 | O75110 | details | | Probable phospholipid-transporting ATPase VD | ATP10D | 4p12 | Q9P241 | details | | Probable phospholipid-transporting ATPase IB | ATP8A2 | 13q12 | Q9NTI2 | details | | Probable phospholipid-transporting ATPase IA | ATP8A1 | 4p13 | Q9Y2Q0 | details | | Probable phospholipid-transporting ATPase IM | ATP8B4 | 15q21.2 | Q8TF62 | details | | Probable phospholipid-transporting ATPase IF | ATP11B | 3q27 | Q9Y2G3 | details | | Probable phospholipid-transporting ATPase IK | ATP8B3 | 19p13.3 | O60423 | details | | 1-acyl-sn-glycerol-3-phosphate acyltransferase gamma | AGPAT3 | 21q22.3 | Q9NRZ7 | details | | 1-acyl-sn-glycerol-3-phosphate acyltransferase beta | AGPAT2 | 9q34.3 | O15120 | details | | 1-acyl-sn-glycerol-3-phosphate acyltransferase alpha | AGPAT1 | 6p21.3 | Q99943 | details | | Phosphatidylinositol-glycan-specific phospholipase D | GPLD1 | 6p22.1 | P80108 | details | | Arf-GAP with GTPase, ANK repeat and PH domain-containing protein 2 | AGAP2 | 12q14.1 | Q99490 | details | | 1-acyl-sn-glycerol-3-phosphate acyltransferase epsilon | AGPAT5 | 8p23.1 | Q9NUQ2 | details | | 2-acylglycerol O-acyltransferase 2 | MOGAT2 | 11q13.5 | Q3SYC2 | details | | Diacylglycerol kinase eta | DGKH | 13q14.11 | Q86XP1 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-3 | PLCD3 | 17q21.31 | Q8N3E9 | details | | 1-acyl-sn-glycerol-3-phosphate acyltransferase delta | AGPAT4 | 6q26 | Q9NRZ5 | details | | Diacylglycerol kinase kappa | DGKK | | Q5KSL6 | details | | Probable phospholipid-transporting ATPase VB | ATP10B | 5q34 | O94823 | details | | Probable phospholipid-transporting ATPase ID | ATP8B2 | 1q21.3 | P98198 | details | | N-acyl-phosphatidylethanolamine-hydrolyzing phospholipase D | NAPEPLD | 7q22.1 | Q6IQ20 | details | | Lysocardiolipin acyltransferase 1 | LCLAT1 | 2p23.1 | Q6UWP7 | details | | Lipase member H | LIPH | 3q27 | Q8WWY8 | details | | Lipase member I | LIPI | 21q11.2 | Q6XZB0 | details | | Lipid phosphate phosphatase-related protein type 4 | LPPR4 | 1p21.2 | Q7Z2D5 | details | | Lipid phosphate phosphatase-related protein type 5 | LPPR5 | 1p21.3 | Q32ZL2 | details | | Presqualene diphosphate phosphatase | PPAPDC2 | 9p24.1 | Q8IY26 | details | | Probable lipid phosphate phosphatase PPAPDC3 | PPAPDC3 | 9q34.13 | Q8NBV4 | details | | Phosphatidate phosphatase PPAPDC1A | PPAPDC1A | 10q26.12 | Q5VZY2 | details | | Phosphatidate phosphatase PPAPDC1B | PPAPDC1B | 8p11.23 | Q8NEB5 | details | | Acylglycerol kinase, mitochondrial | AGK | 7q34 | Q53H12 | details | | Phospholipase D1 variant | | | Q59EA4 | details | | 1-acylglycerol-3-phosphate O-acyltransferase 6 (Lysophosphatidic acid acyltransferase, zeta), isoform CRA_b | AGPAT6 | 8p11.21 | Q2TU73 | details | | Diacylglycerol kinase, epsilon 64kDa | DGKE | 17q22 | A1L4Q0 | details | | Arf-GAP with GTPase, ANK repeat and PH domain-containing protein 1 | AGAP1 | 2q37 | Q9UPQ3 | details | | Phospholipase DDHD2 | DDHD2 | 8p11.23 | O94830 | details |
|
|---|
| Transporters | |
| Probable phospholipid-transporting ATPase IG | ATP11C | | Q8NB49 | details | | Probable phospholipid-transporting ATPase IH | ATP11A | 13q34 | P98196 | details | | Probable phospholipid-transporting ATPase VA | ATP10A | 15q11.2 | O60312 | details | | Probable phospholipid-transporting ATPase IC | ATP8B1 | 18q21-q22|18q21.31 | O43520 | details | | Probable phospholipid-transporting ATPase IIA | ATP9A | 20q13.2 | O75110 | details | | Probable phospholipid-transporting ATPase VD | ATP10D | 4p12 | Q9P241 | details | | Probable phospholipid-transporting ATPase IB | ATP8A2 | 13q12 | Q9NTI2 | details | | Probable phospholipid-transporting ATPase IA | ATP8A1 | 4p13 | Q9Y2Q0 | details | | Probable phospholipid-transporting ATPase IM | ATP8B4 | 15q21.2 | Q8TF62 | details | | Probable phospholipid-transporting ATPase IF | ATP11B | 3q27 | Q9Y2G3 | details | | Probable phospholipid-transporting ATPase IK | ATP8B3 | 19p13.3 | O60423 | details | | Probable phospholipid-transporting ATPase VB | ATP10B | 5q34 | O94823 | details | | Probable phospholipid-transporting ATPase ID | ATP8B2 | 1q21.3 | P98198 | details |
|
|---|
| Metal Bindings | |
| Diacylglycerol kinase gamma | DGKG | 3q27.2-q27.3 | P49619 | details | | Diacylglycerol kinase alpha | DGKA | 12q13.3 | P23743 | details | | Diacylglycerol kinase beta | DGKB | 7p21.2 | Q9Y6T7 | details | | Phospholipase DDHD1 | DDHD1 | 14q21 | Q8NEL9 | details | | cAMP-specific 3',5'-cyclic phosphodiesterase 4D | PDE4D | 5q12 | Q08499 | details | | Probable phospholipid-transporting ATPase IG | ATP11C | | Q8NB49 | details | | Probable phospholipid-transporting ATPase IH | ATP11A | 13q34 | P98196 | details | | Probable phospholipid-transporting ATPase VA | ATP10A | 15q11.2 | O60312 | details | | Probable phospholipid-transporting ATPase IC | ATP8B1 | 18q21-q22|18q21.31 | O43520 | details | | Probable phospholipid-transporting ATPase IIA | ATP9A | 20q13.2 | O75110 | details | | Probable phospholipid-transporting ATPase VD | ATP10D | 4p12 | Q9P241 | details | | Probable phospholipid-transporting ATPase IB | ATP8A2 | 13q12 | Q9NTI2 | details | | Probable phospholipid-transporting ATPase IA | ATP8A1 | 4p13 | Q9Y2Q0 | details | | Probable phospholipid-transporting ATPase IM | ATP8B4 | 15q21.2 | Q8TF62 | details | | Probable phospholipid-transporting ATPase IF | ATP11B | 3q27 | Q9Y2G3 | details | | Probable phospholipid-transporting ATPase IK | ATP8B3 | 19p13.3 | O60423 | details | | Arf-GAP with GTPase, ANK repeat and PH domain-containing protein 2 | AGAP2 | 12q14.1 | Q99490 | details | | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-3 | PLCD3 | 17q21.31 | Q8N3E9 | details | | Probable phospholipid-transporting ATPase VB | ATP10B | 5q34 | O94823 | details | | Probable phospholipid-transporting ATPase ID | ATP8B2 | 1q21.3 | P98198 | details | | N-acyl-phosphatidylethanolamine-hydrolyzing phospholipase D | NAPEPLD | 7q22.1 | Q6IQ20 | details | | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1 | ACAP1 | 17p13.1 | Q15027 | details | | Arf-GAP with GTPase, ANK repeat and PH domain-containing protein 1 | AGAP1 | 2q37 | Q9UPQ3 | details | | Phospholipase DDHD2 | DDHD2 | 8p11.23 | O94830 | details | | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 2 | ACAP2 | 3q29 | Q15057 | details |
|
|---|
| Receptors | |
|---|
| Transcriptional Factors | |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000674 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB022175 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 393518 |
|---|
| KEGG Compound ID | C00416 |
|---|
| BioCyc ID | CPD0-1422 |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| METLIN ID | 5644 |
|---|
| PubChem Compound | 446066 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 73246 |
|---|
| References |
|---|
| General References | Not Available |
|---|

