| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:26:12 UTC |
|---|
| Updated at | 2020-11-18 16:34:50 UTC |
|---|
| CannabisDB ID | CDB000138 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | D-Mannose |
|---|
| Description | Mannose or D-Mannose, also known as mannopyranose or carubinose, is a C-2 epimer of glucose. It belongs to the class of organic compounds known as hexoses. These are monosaccharides in which the sugar unit is a six-carbon containing moeity. Mannose contains an aldehyde group and is therefore referred to as an aldohexose. Mannose commonly exists with two different-sized rings, the pyranose (six-membered) form and the furanose (five-membered) form. Each ring closure can have either an alpha or beta configuration at the anomeric position. Therefore, mannose exists in four different forms. The α‐form of D‐Mannose is mild and sweet, whereas the β‐form is bitter. Hence, D‐mannose is sweet with a bitter aftertaste. D-mannose is 0.6 times as sweet as sucrose. Sweetness is detected through the binding of sugars to the T1R3 and T1R2 proteins, to form a G-protein coupled receptor that is the sweetness receptor in mammals. D-Mannose is a hydrophilic, neutral molecule that is highly water soluble. It exists as a white crystalline powder. Mannose can be formed by the oxidation of mannitol. Because mannose is a glucose epimer, it can also be naturally produced in a wide range of organisms from glucose or fructose using D-mannose isomerases or D-xylose isomerases. The origin of the name mannose comes from the Hebrew word “manna” or food ( Ref:DOI > Ref:DOI > Ref:DOI ). Several trees and shrubs can produce a substance called manna, such as the “manna tree” from whose secretions mannitol was originally isolated. Mannose is an important component of polysaccharides and glycoproteins. Mannose is the dominant monosaccharide in the N-linked glycosylation of proteins in both plants and animals. Several congenital disorders of glycosylation are associated with mutations in enzymes involved in mannose metabolism (PMID: 20363348 ). Normally in those mammalian proteins that are glycosylated, the mannose moieties are shielded by galactose, sialic acid or GlcNAc groups. This is important, as the innate immune system in mammals is geared to recognise exposed mannose residues. Many yeasts and viruses (including the AIDS virus) are decorated with exposed mannans or mannose residues, which generates a strong innate immune response. In particular, Mannose-binding lectin (MBL) is an important constituent of the innate immune system. This protein binds through multiple lectin domains to the repeating sugar arrays that decorate many microbial surfaces and is then able to activate the complement system through a specific protease called MBL-associated protease-2. The digestion of many polysaccharides and glycoproteins by humans and other organisms yields mannose, which is phosphorylated by hexokinase to generate mannose-6-phosphate. Mannose-6-phosphate is then converted to fructose-6-phosphate, by the enzyme phosphomannose isomerase, and then enters the glycolytic pathway. The primary pathway for the formation of L-fucose in prokaryotic and eukaryotic cells is from D-mannose via an internal oxidation-reduction and then epimerization of GDP-D-mannose to produce GDP-L-fucose (PMID: 9488699 , 16154739 , 11414367 ). D-Mannose or Mannose is found in many plants or plant foods where it occurs as a component of mannan, hemicellulose, or cellulose in various cell wall architectures. Individual D-Mannose units can be generated from the decomposition of hemicelluloses by extracellular xylanases. The content of D-mannose reaches up to 21.2% dry weight (DW) in spent coffee grounds. Likewise, jujube powder has up to 12.9% D-mannose. Other plants or plant components with high levels of Mannose include orange peel, litchi pericarp, cranberries, apples, mangoes and peaches. Soybean, kidney bean, sword bean, peanut, and other leguminous plants contain plenty of lectin, which is a glycoprotein containing 3.8% neutral carbohydrates including D-mannose. D-Mannose is also found in Cannabis plants (PMID: 6991645 ). Mannose (D-mannose) can be used as a nutritional supplement to prevent recurrent urinary tract infections. D-Mannose appears to stop certain bacteria from sticking to the bladder walls by blocking the interaction of the FimH protein on bacterial pili (PMID: 23633128 ). D-Mannose has been widely used in the food, pharmaceutical, and poultry industries, acting as the source of dietary supplements, starting material for the synthesis of drugs and blocking colonization in animal feeds ( Ref:DOI > Ref:DOI > Ref:DOI ). D-Mannose is able to stimulate insulin secretion. The α‐anomer of D‐mannose is better able than the β‐anomer to stimulate insulin release. D‐Mannose is well metabolized in pancreatic islets of Langerhans and is known to penetrate the blood–brain barrier. When mannan is incorporated into a diet, a number of interesting effects are observed, including a decrease in appetite and a reduction in S‐glucose and cholesterol absorption ( Ref:DOI > Ref:DOI > Ref:DOI ) |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| alpha-D-Man | ChEBI | | a-D-Man | Generator | | α-D-Man | Generator | | a-D-Mannose | Generator | | α-D-Mannose | Generator | | alpha-D-Mannopyranose | ChEBI | | a-D-Mannopyranose | Generator | | α-D-Mannopyranose | Generator | | (+)-Mannose | HMDB | | Carubinose | HMDB | | D(+)-Mannose | HMDB | | Mannose | HMDB | | Seminose | HMDB | | alpha-D-Mannose | HMDB | | alpha-Mannose | HMDB | | α-Mannose | HMDB |
|
|---|
| Chemical Formula | C6H12O6 |
|---|
| Average Molecular Weight | 180.16 |
|---|
| Monoisotopic Molecular Weight | 180.0634 |
|---|
| IUPAC Name | (2S,3S,4S,5S,6R)-6-(hydroxymethyl)oxane-2,3,4,5-tetrol |
|---|
| Traditional Name | α-D-mannose |
|---|
| CAS Registry Number | 29696-75-1 |
|---|
| SMILES | OC[C@H]1O[C@H](O)[C@@H](O)[C@@H](O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h2-11H,1H2/t2-,3-,4+,5+,6+/m1/s1 |
|---|
| InChI Key | WQZGKKKJIJFFOK-PQMKYFCFSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as hexoses. These are monosaccharides in which the sugar unit is a is a six-carbon containing moeity. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Hexoses |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hexose monosaccharide
- Oxane
- Secondary alcohol
- Hemiacetal
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Hydrocarbon derivative
- Primary alcohol
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 132 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 713 mg/mL at 17 °C | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | D-Mannose, 5 TMS, GC-MS Spectrum | splash10-0mi2-0931000000-96f0cf3c2d4a8bea25b9 | Spectrum | | GC-MS | D-Mannose, 5 TMS, GC-MS Spectrum | splash10-00kb-1931000000-ec423f92582012821c6a | Spectrum | | GC-MS | D-Mannose, 5 TMS, GC-MS Spectrum | splash10-0fr2-1920000000-b6be17dc79a948dfe5fa | Spectrum | | GC-MS | D-Mannose, 5 TMS; 1 MEOX, GC-MS Spectrum | splash10-00di-9831000000-409527d22fb759cb628f | Spectrum | | GC-MS | D-Mannose, 5 TMS; 1 MEOX, GC-MS Spectrum | splash10-00di-9621000000-34b421fd962840bfbaec | Spectrum | | GC-MS | D-Mannose, 1 MEOX; 5 TMS, GC-MS Spectrum | splash10-066r-1942000000-1c8164b54d2d16c782c3 | Spectrum | | GC-MS | D-Mannose, 1 MEOX; 5 TMS, GC-MS Spectrum | splash10-0ldi-1942000000-c68a088eba727f3a4ada | Spectrum | | GC-MS | D-Mannose, non-derivatized, GC-MS Spectrum | splash10-0mi2-0931000000-96f0cf3c2d4a8bea25b9 | Spectrum | | GC-MS | D-Mannose, non-derivatized, GC-MS Spectrum | splash10-00kb-1931000000-ec423f92582012821c6a | Spectrum | | GC-MS | D-Mannose, non-derivatized, GC-MS Spectrum | splash10-0fr2-1920000000-b6be17dc79a948dfe5fa | Spectrum | | GC-MS | D-Mannose, non-derivatized, GC-MS Spectrum | splash10-00di-9831000000-409527d22fb759cb628f | Spectrum | | GC-MS | D-Mannose, non-derivatized, GC-MS Spectrum | splash10-00di-9621000000-34b421fd962840bfbaec | Spectrum | | GC-MS | D-Mannose, non-derivatized, GC-MS Spectrum | splash10-066r-1942000000-1c8164b54d2d16c782c3 | Spectrum | | GC-MS | D-Mannose, non-derivatized, GC-MS Spectrum | splash10-0ldi-1942000000-c68a088eba727f3a4ada | Spectrum | | Predicted GC-MS | D-Mannose, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0np0-9700000000-e8d638dc817e46b97d7b | Spectrum | | Predicted GC-MS | D-Mannose, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | D-Mannose, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | D-Mannose, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | D-Mannose, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | D-Mannose, TMS_1_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | D-Mannose, TMS_1_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | D-Mannose, TMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | D-Mannose, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | D-Mannose, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | D-Mannose, TMS_2_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-01pc-6900000000-0ef33e32ee16b7803f1b | 2020-03-26 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-00kr-9000000000-27e169c1df0c3ee59806 | 2020-03-26 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-03ea-0900000000-9fb0e8db39598c7a269b | 2020-03-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01q9-0900000000-b0bc47623e7b2ca31c02 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03ea-3900000000-648e1637af29cf2a3518 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0007-9200000000-9e6f46a1cbf52d6e347a | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-2900000000-a4ec4f0b1e29e360a952 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01t9-6900000000-7b3ea9c64ecc8d4ac867 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9100000000-ec2bf4918640a0a36398 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-2900000000-2448926b508622464fe7 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-056r-8900000000-4c073cb93b78120113e6 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-ece70093ab5d3c331ac4 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01qa-0900000000-04ceb34d441ff6a75763 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dl-9400000000-a8f1ceab155611f949c1 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01ow-9000000000-358f68fc2b7a72c27546 | 2021-09-22 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 22.53 MHz, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Fructose and Mannose Degradation | 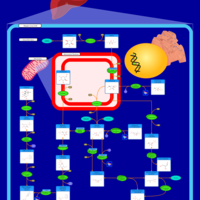 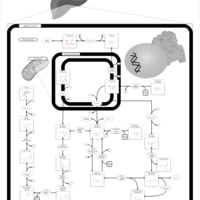 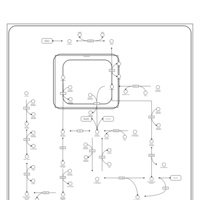 |  | | Galactose Metabolism | 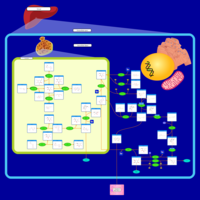 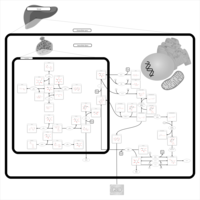 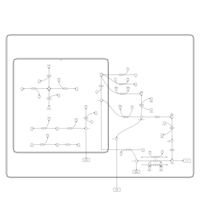 |  | | Fructosuria | 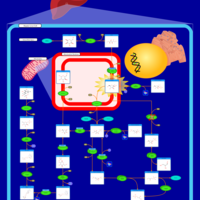 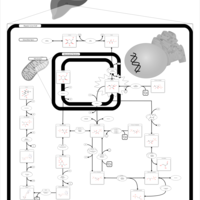 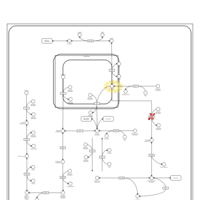 | Not Available | | Fructose intolerance, hereditary | 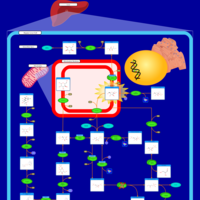 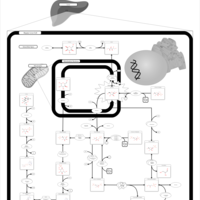 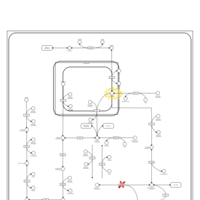 | Not Available | | Galactosemia | 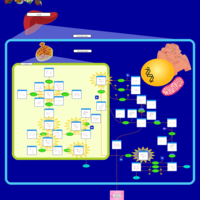 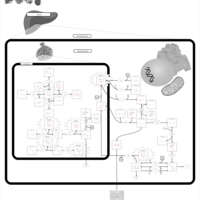 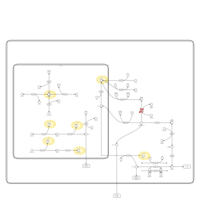 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
| Cation-independent mannose-6-phosphate receptor | IGF2R | 6q26 | P11717 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | |
| Phospholipase A2 | PLA2G1B | 12q23-q24.1 | P04054 | details | | Nitric oxide synthase, brain | NOS1 | 12q24.2-q24.31 | P29475 | details | | N-acetylglucosamine-1-phosphotransferase subunits alpha/beta | GNPTAB | 12q23.2 | Q3T906 | details |
|
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000169 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB001202 |
|---|
| KNApSAcK ID | C00001126 |
|---|
| Chemspider ID | 161434 |
|---|
| KEGG Compound ID | C00936 |
|---|
| BioCyc ID | MANNOSE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Mannose |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 185698 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 28729 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
- Ito Y, Hagihara S, Matsuo I, Totani K: Structural approaches to the study of oligosaccharides in glycoprotein quality control. Curr Opin Struct Biol. 2005 Oct;15(5):481-9. doi: 10.1016/j.sbi.2005.08.012. [PubMed:16154739 ]
- Yoshida Y: Lectin-Type Ubiquitin Ligase Subunits: Fbs Proteins and Their Applications for Use. Methods Mol Biol. 2020;2132:215-224. doi: 10.1007/978-1-0716-0430-4_22. [PubMed:32306330 ]
- Freeze HH, Sharma V: Metabolic manipulation of glycosylation disorders in humans and animal models. Semin Cell Dev Biol. 2010 Aug;21(6):655-62. doi: 10.1016/j.semcdb.2010.03.011. Epub 2010 Apr 2. [PubMed:20363348 ]
- Park SH, Pastuszak I, Drake R, Elbein AD: Purification to apparent homogeneity and properties of pig kidney L-fucose kinase. J Biol Chem. 1998 Mar 6;273(10):5685-91. doi: 10.1074/jbc.273.10.5685. [PubMed:9488699 ]
- Jack DL, Klein NJ, Turner MW: Mannose-binding lectin: targeting the microbial world for complement attack and opsonophagocytosis. Immunol Rev. 2001 Apr;180:86-99. doi: 10.1034/j.1600-065x.2001.1800108.x. [PubMed:11414367 ]
- Kranjcec B, Papes D, Altarac S: D-mannose powder for prophylaxis of recurrent urinary tract infections in women: a randomized clinical trial. World J Urol. 2014 Feb;32(1):79-84. doi: 10.1007/s00345-013-1091-6. Epub 2013 Apr 30. [PubMed:23633128 ]
|
|---|

