| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:25:56 UTC |
|---|
| Updated at | 2022-12-13 23:36:22 UTC |
|---|
| CannabisDB ID | CDB000131 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | L-Serine |
|---|
| Description | Serine (or L-serine), abbreviated as Ser or S, is a nonessential amino acid derived from glycine. As all amino acid are building blocks of protein and peptides, serine can become essential under certain conditions, and is thus important in maintaining health and preventing disease. Muscles have low-average concentration of serine compared to other amino acids. . In contrast, serine is highly concentrated in all cell membranes. L-Serine may be derived from four possible sources: dietary intake, biosynthesis from the glycolytic intermediate 3-phosphoglycerate, from glycine, and by protein and phospholipid degradation. Little data is available on the relative contributions of each of these four sources of L-serine to serine homoeostasis. It is likely that the predominant source of L-serine varies in different tissues and during different stages of human development. In the biosynthetic pathway, the glycolytic intermediate 3-phosphoglycerate is converted to serine by a three enzyme cascade. First, 3-phosphoglycerate is converted into phosphohydroxypyruvate by 3-phosphoglycerate dehydrogenase (3- PGDH; EC 1.1.1.95). Second, phosphohydroxypyruvate is metabolized to phosphoserine by phosphohydroxypyruvate aminotransferase (EC 2.6.1.52) and third, phosphoserine is converted into l-serine by phosphoserine phosphatase (PSP; EC 3.1.3.3). In liver tissue, the serine biosynthetic pathway is regulated in response to dietary and hormonal changes. Of the three biosynthetic enzymes, the properties of 3-PGDH and PSP are the best documented. Hormonal factors such as glucagon and corticosteroids also influence 3-PGDH and PSP activities in interactions dependent upon the diet. L-serine plays a central role in cellular proliferation. L-Serine is the predominant source of one-carbon groups for the de novo synthesis of purine nucleotides and deoxythymidine monophosphate. It has long been recognized that, in cell cultures, L-serine is a conditional essential amino acid, because it cannot be synthesized in sufficient quantities to meet the cellular demands for its utilization. In recent years, L-serine and the products of its metabolism have been recognized not only to be essential for cell proliferation, but also to be necessary for specific functions in the central nervous system. The findings of altered levels of serine and glycine in patients with psychiatric disorders and the severe neurological abnormalities in patients with defects of L-serine synthesis underscore the importance of L-serine in brain development and function (PMID: 12534373 ). L-serine is also found in Cannabis plants (PMID: 6991645 ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S)-2-Amino-3-hydroxypropanoic acid | ChEBI | | (S)-(-)-Serine | ChEBI | | (S)-2-Amino-3-hydroxypropanoic acid | ChEBI | | (S)-alpha-Amino-beta-hydroxypropionic acid | ChEBI | | (S)-Serine | ChEBI | | beta-Hydroxy-L-alanine | ChEBI | | beta-Hydroxyalanine | ChEBI | | L-(-)-Serine | ChEBI | | L-2-Amino-3-hydroxypropionic acid | ChEBI | | L-3-Hydroxy-2-aminopropionic acid | ChEBI | | L-3-Hydroxy-alanine | ChEBI | | L-Ser | ChEBI | | L-Serin | ChEBI | | S | ChEBI | | Ser | ChEBI | | SERINE | ChEBI | | (2S)-2-Amino-3-hydroxypropanoate | Generator | | (S)-2-Amino-3-hydroxypropanoate | Generator | | (S)-a-Amino-b-hydroxypropionate | Generator | | (S)-a-Amino-b-hydroxypropionic acid | Generator | | (S)-alpha-Amino-beta-hydroxypropionate | Generator | | (S)-Α-amino-β-hydroxypropionate | Generator | | (S)-Α-amino-β-hydroxypropionic acid | Generator | | b-Hydroxy-L-alanine | Generator | | Β-hydroxy-L-alanine | Generator | | b-Hydroxyalanine | Generator | | Β-hydroxyalanine | Generator | | L-2-Amino-3-hydroxypropionate | Generator | | L-3-Hydroxy-2-aminopropionate | Generator | | (-)-Serine | HMDB | | (S)-2-Amino-3-hydroxy-propanoate | HMDB | | (S)-2-Amino-3-hydroxy-propanoic acid | HMDB | | (S)-b-Amino-3-hydroxypropionate | HMDB | | (S)-b-Amino-3-hydroxypropionic acid | HMDB | | (S)-beta-Amino-3-hydroxypropionate | HMDB | | (S)-beta-Amino-3-hydroxypropionic acid | HMDB | | 2-Amino-3-hydroxypropanoate | HMDB | | 2-Amino-3-hydroxypropanoic acid | HMDB | | 3-Hydroxy-L-alanine | HMDB | | L Serine | HMDB | | (3R,3'r,6S)-4,5-DIDEHYDRO-5,6-dihydro-BETA,BETA-carotene-3,3'-diol | HMDB | | Bo-xan | HMDB | | e 161b | HMDB | | Xanthophyll | HMDB | | (3R,3'r,6S)-4,5-DIDEHYDRO-5,6-dihydro-b,b-carotene-3,3'-diol | HMDB | | (3R,3'r,6S)-4,5-DIDEHYDRO-5,6-dihydro-β,β-carotene-3,3'-diol | HMDB |
|
|---|
| Chemical Formula | C3H7NO3 |
|---|
| Average Molecular Weight | 105.09 |
|---|
| Monoisotopic Molecular Weight | 105.0426 |
|---|
| IUPAC Name | (2S)-2-amino-3-hydroxypropanoic acid |
|---|
| Traditional Name | L-serine |
|---|
| CAS Registry Number | 6898-95-9 |
|---|
| SMILES | N[C@@H](CO)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C3H7NO3/c4-2(1-5)3(6)7/h2,5H,1,4H2,(H,6,7)/t2-/m0/s1 |
|---|
| InChI Key | MTCFGRXMJLQNBG-REOHCLBHSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as serine and derivatives. Serine and derivatives are compounds containing serine or a derivative thereof resulting from reaction of serine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Serine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Serine or derivatives
- Alpha-amino acid
- L-alpha-amino acid
- Beta-hydroxy acid
- Hydroxy acid
- Amino acid
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Alcohol
- Primary amine
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Hydrocarbon derivative
- Primary aliphatic amine
- Organic oxide
- Organopnictogen compound
- Carbonyl group
- Organic oxygen compound
- Amine
- Organic nitrogen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 228 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 425 mg/mL | Not Available | | logP | -3.07 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-0200-9000000000-dc5e2afcfab602641180 | 2018-05-25 | View Spectrum | | GC-MS | L-Serine, 3 TMS, GC-MS Spectrum | splash10-0udi-0940000000-59d5e0f76fd204be8110 | Spectrum | | GC-MS | L-Serine, 3 TMS, GC-MS Spectrum | splash10-0udi-0980000000-ab49fc3ee88db87d1a16 | Spectrum | | GC-MS | L-Serine, 2 TMS, GC-MS Spectrum | splash10-0159-0900000000-8e169037bcbe1dcd5de8 | Spectrum | | GC-MS | L-Serine, 3 TMS, GC-MS Spectrum | splash10-0uxr-0970000000-67271ed235e35c662db5 | Spectrum | | GC-MS | L-Serine, 3 TMS, GC-MS Spectrum | splash10-00di-9540000000-1297bf302358bcf0f1c0 | Spectrum | | GC-MS | L-Serine, 3 TMS, GC-MS Spectrum | splash10-0uxr-1790000000-caaa37778f5a4686f5f0 | Spectrum | | GC-MS | L-Serine, non-derivatized, GC-MS Spectrum | splash10-0159-0900000000-ae4f860052c41e9ba734 | Spectrum | | GC-MS | L-Serine, non-derivatized, GC-MS Spectrum | splash10-0uxr-0590000000-ce32a8bfb64d16590b52 | Spectrum | | GC-MS | L-Serine, non-derivatized, GC-MS Spectrum | splash10-0udi-0940000000-59d5e0f76fd204be8110 | Spectrum | | GC-MS | L-Serine, non-derivatized, GC-MS Spectrum | splash10-0udi-0980000000-ab49fc3ee88db87d1a16 | Spectrum | | GC-MS | L-Serine, non-derivatized, GC-MS Spectrum | splash10-0159-0900000000-8e169037bcbe1dcd5de8 | Spectrum | | GC-MS | L-Serine, non-derivatized, GC-MS Spectrum | splash10-0uxr-0970000000-67271ed235e35c662db5 | Spectrum | | GC-MS | L-Serine, non-derivatized, GC-MS Spectrum | splash10-00dr-7966000000-7b5b37fd4ac885548172 | Spectrum | | GC-MS | L-Serine, non-derivatized, GC-MS Spectrum | splash10-00di-9540000000-1297bf302358bcf0f1c0 | Spectrum | | GC-MS | L-Serine, non-derivatized, GC-MS Spectrum | splash10-0uxr-1790000000-caaa37778f5a4686f5f0 | Spectrum | | Predicted GC-MS | L-Serine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-06sl-9000000000-033631650075b940e0e2 | Spectrum | | Predicted GC-MS | L-Serine, 2 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0fkl-9420000000-ed7a327e1f071802a4dd | Spectrum | | Predicted GC-MS | L-Serine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Serine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Serine, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Serine, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Serine, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Serine, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Serine, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Serine, TMS_2_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-03di-9100000000-38f84912ebfc7b4e8339 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-03dl-9000000000-89e57995d3ca765a20e8 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-01ox-9100000000-f3baf93032cc9f09cf2d | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0zfr-0900000000-b6085e53e881cdaa1824 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-2e16e9d6f9b3c2688f8a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0a4i-9000000000-9857daa028fb3016a7e2 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0006-0009200000-0617acd25c69bf24ad1f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0a4i-0900000000-823dfcf6229956372e31 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0a4i-9000000000-eec63565f8a9fe4bd336 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-008412efd202c202d56a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0002-0930000000-932aedc4efb5e1dd5e7b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0w29-0951000000-cd0895e475dd50e4934d | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-9000000000-15294ba22b153cf5ab5b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-014i-9000000000-48c370acff657443f0e6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0udi-0190000000-1bad10ba7388ce0c2249 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0ue9-0792100000-9f2bfe40e4c01044d209 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-9000000000-dfa7aca58cc28303ebc7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0udi-0900000000-282f31d8021649afdbe8 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0udi-0290000000-eeee98e6b0514b08cd63 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0udi-1900000000-ed66566e27a94ee99f13 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-00di-9100000000-b9f6b4b6ee7c88a9c569 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-00di-9000000000-929aea63ace0e20c6d43 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-00di-9000000000-ba9cd5b3e37ded5d3764 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0a4i-6900000000-5622633ab0a27d2e5360 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-03di-9000000000-3e25ca80e2e62be19ac5 | 2012-08-31 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Ammonia Recycling |  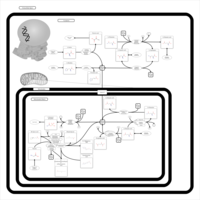 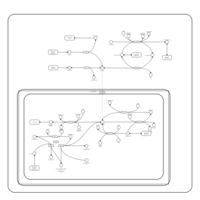 |  | | Sphingolipid Metabolism | 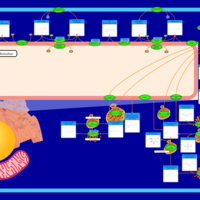 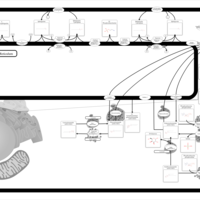 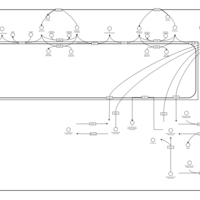 |  | | Glycine and Serine Metabolism | 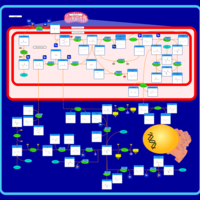 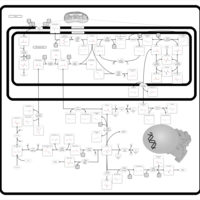 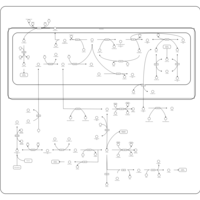 |  | | Methionine Metabolism |    |  | | Homocysteine Degradation |   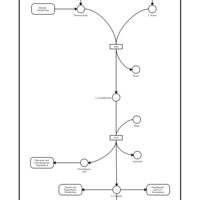 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 0.637 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 0.372 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 0.511 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.404 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 0.386 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 0.587 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000187 |
|---|
| DrugBank ID | DB00133 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB012739 |
|---|
| KNApSAcK ID | C00001393 |
|---|
| Chemspider ID | 5736 |
|---|
| KEGG Compound ID | C00065 |
|---|
| BioCyc ID | SER |
|---|
| BiGG ID | 33717 |
|---|
| Wikipedia Link | L-serine |
|---|
| METLIN ID | 5203 |
|---|
| PubChem Compound | 5951 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17115 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
- de Koning TJ, Snell K, Duran M, Berger R, Poll-The BT, Surtees R: L-serine in disease and development. Biochem J. 2003 May 1;371(Pt 3):653-61. doi: 10.1042/BJ20021785. [PubMed:12534373 ]
|
|---|



