| Identification |
|---|
| HMDB Protein ID
| CDBP04181 |
| Secondary Accession Numbers
| Not Available |
| Name
| Serine racemase |
| Description
| Not Available |
| Synonyms
|
- D-serine ammonia-lyase
- D-serine dehydratase
- L-serine ammonia-lyase
- L-serine dehydratase
|
| Gene Name
| SRR |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in catalytic activity |
| Specific Function
| Catalyzes the synthesis of D-serine from L-serine. D-serine is a key coagonist with glutamate at NMDA receptors. Has dehydratase activity towards both L-serine and D-serine.
|
| GO Classification
|
| Biological Process |
| response to lipopolysaccharide |
| response to drug |
| D-serine biosynthetic process |
| L-serine metabolic process |
| brain development |
| aging |
| protein homotetramerization |
| response to morphine |
| pyruvate biosynthetic process |
| Cellular Component |
| apical part of cell |
| cytoplasm |
| plasma membrane |
| neuronal cell body |
| Function |
| binding |
| catalytic activity |
| cofactor binding |
| pyridoxal phosphate binding |
| Molecular Function |
| calcium ion binding |
| glycine binding |
| magnesium ion binding |
| ATP binding |
| D-serine ammonia-lyase activity |
| serine racemase activity |
| threonine racemase activity |
| pyridoxal phosphate binding |
| protein homodimerization activity |
| L-serine ammonia-lyase activity |
| Process |
| metabolic process |
| cellular metabolic process |
| cellular amino acid and derivative metabolic process |
| cellular amino acid metabolic process |
|
| Cellular Location
|
Not Available
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Glycine and Serine Metabolism | 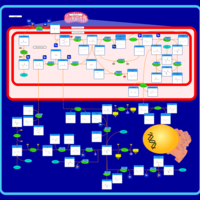 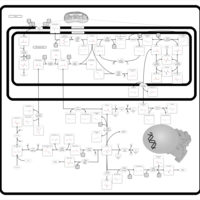 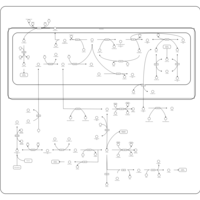 |  | | Glycine, serine and threonine metabolism | Not Available |  | | Dimethylglycine Dehydrogenase Deficiency | 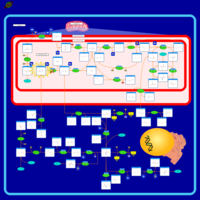   | Not Available | | Dihydropyrimidine Dehydrogenase Deficiency (DHPD) |    | Not Available | | Sarcosinemia | 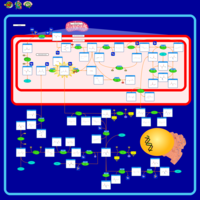 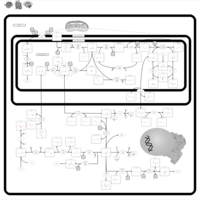 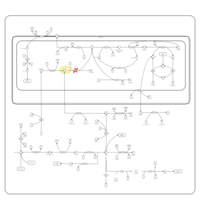 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 17 |
| Locus
| 17p13 |
| SNPs
| SRR |
| Gene Sequence
|
>1023 bp
ATGTGTGCTCAGTATTGCATCTCCTTTGCTGATGTTGAAAAAGCTCATATCAACATTCGA
GATTCTATCCACCTCACACCAGTGCTAACAAGCTCCATTTTGAATCAACTAACAGGGCGC
AATCTTTTCTTCAAATGTGAACTCTTCCAGAAAACAGGATCTTTTAAGATTCGTGGTGCT
CTCAATGCCGTCAGAAGCTTGGTTCCTGATGCTTTAGAAAGGAAGCCGAAAGCTGTTGTT
ACTCACAGCAGTGGAAACCATGGCCAGGCTCTCACCTATGCTGCCAAATTGGAAGGAATT
CCTGCTTATATTGTGGTGCCCCAGACAGCTCCAGACTGTAAAAAACTTGCAATACAAGCC
TACGGAGCGTCAATTGTATACTGTGAACCTAGTGATGAGTCCAGAGAAAATGTTGCAAAA
AGAGTTACAGAAGAAACAGAAGGCATCATGGTACATCCCAACCAGGAGCCTGCAGTGATA
GCTGGACAAGGGACAATTGCCCTGGAAGTGCTGAACCAGGTTCCTTTGGTGGATGCACTG
GTGGTACCTGTAGGTGGAGGAGGAATGCTTGCTGGAATAGCAATTACAGTTAAGGCTCTG
AAACCTAGTGTGAAGGTATATGCTGCTGAACCCTCAAATGCAGATGACTGCTACCAGTCC
AAGCTGAAGGGGAAACTGATGCCCAATCTTTATCCTCCAGAAACCATAGCAGATGGTGTC
AAATCCAGCATTGGCTTGAACACCTGGCCTATTATCAGGGACCTTGTGGATGATATCTTC
ACTGTCACAGAGGATGAAATTAAGTGTGCAACCCAGCTGGTGTGGGAGAGGATGAAACTA
CTCATTGAACCTACAGCTGGTGTTGGAGTGGCTGCTGTGCTGTCTCAACATTTTCAAACT
GTTTCCCCAGAAGTAAAGAACATTTGTATTGTGCTCAGTGGTGGAAATGTAGACTTAACC
TCCTCCATAACTTGGGTGAAGCAGGCTGAAAGGCCAGCTTCTTATCAGTCTGTTTCTGTT
TAA
|
| Protein Properties |
|---|
| Number of Residues
| 340 |
| Molecular Weight
| 36565.905 |
| Theoretical pI
| 6.534 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Serine racemase
MCAQYCISFADVEKAHINIRDSIHLTPVLTSSILNQLTGRNLFFKCELFQKTGSFKIRGA
LNAVRSLVPDALERKPKAVVTHSSGNHGQALTYAAKLEGIPAYIVVPQTAPDCKKLAIQA
YGASIVYCEPSDESRENVAKRVTEETEGIMVHPNQEPAVIAGQGTIALEVLNQVPLVDAL
VVPVGGGGMLAGIAITVKALKPSVKVYAAEPSNADDCYQSKLKGKLMPNLYPPETIADGV
KSSIGLNTWPIIRDLVDDIFTVTEDEIKCATQLVWERMKLLIEPTAGVGVAAVLSQHFQT
VSPEVKNICIVLSGGNVDLTSSITWVKQAERPASYQSVSV
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| Q9GZT4 |
| UniProtKB/Swiss-Prot Entry Name
| SRR_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AF169974 |
| GeneCard ID
| SRR |
| GenAtlas ID
| SRR |
| HGNC ID
| HGNC:14398 |
| References |
|---|
| General References
| Not Available |
