| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:10:48 UTC |
|---|
| Updated at | 2020-12-07 19:11:37 UTC |
|---|
| CannabisDB ID | CDB005115 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Tetrahydrofolic acid |
|---|
| Description | Tetrahydrofolic acid, also known as tetrahydrofolate, belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. Tetrahydrofolic acid is a moderately basic compound (based on its pKa). Tetrahydrofolic acid exists in all eukaryotes, ranging from yeast to humans. Many bacteria use dihydropteroate synthetase to produce dihydropteroate, a molecule without function in humans. Within humans, tetrahydrofolic acid participates in a number of enzymatic reactions. In particular, tetrahydrofolic acid can be converted into dihydrofolic acid; which is mediated by the enzyme dihydrofolate reductase. In addition, tetrahydrofolic acid and formic acid can be converted into 10-formyltetrahydrofolate; which is mediated by the enzyme C-1-tetrahydrofolate synthase, cytoplasmic. In humans, tetrahydrofolic acid is involved in the metabolic disorder called the methylenetetrahydrofolate reductase deficiency (mthfrd) pathway. 10-formyltetrahydrofolate acts as a donor of a group with one carbon atom. Methotrexate acts on dihydrofolate reductase, like pyrimethamine or trimethoprim, as an inhibitor and thus reduces the amount of tetrahydrofolate made. Tetrahydrofolic acid is involved in the conversion of formiminoglutamic acid to glutamic acid; this may reduce the amount of histidine available for decarboxylation and protein synthesis, and hence the urinary histamine and formiminoglutamic acid may be decreased. Tetrahydrofolic acid (THFA), or tetrahydrofolate, is a folic acid derivative. Tetrahydrofolic acid is a cofactor in many reactions, especially in the synthesis (or anabolism) of amino acids and nucleic acids. This may result in megaloblastic anemia. A shortage in tetrahydrofolic acid (FH4) can cause megaloblastic anemia. Tetrahydrofolic acid is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Tetrahydrofolate | Generator | | (6S)-Tetrahydrofolate | HMDB | | (6S)-Tetrahydrofolic acid | HMDB | | 5,6,7,8-Tetrahydrofolate | HMDB | | 5,6,7,8-Tetrahydrofolic acid | HMDB | | Tetra-H-folate | HMDB | | Tetrahydrafolate | HMDB | | Tetrahydropteroyl mono-L-glutamate | HMDB | | Tetrahydropteroylglutamate | HMDB |
|
|---|
| Chemical Formula | C19H23N7O6 |
|---|
| Average Molecular Weight | 445.43 |
|---|
| Monoisotopic Molecular Weight | 445.171 |
|---|
| IUPAC Name | 2-{[4-({[(6S)-4-hydroxy-2-imino-1,2,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]formamido}pentanedioic acid |
|---|
| Traditional Name | 2-{[4-({[(6S)-4-hydroxy-2-imino-5,6,7,8-tetrahydro-1H-pteridin-6-yl]methyl}amino)phenyl]formamido}pentanedioic acid |
|---|
| CAS Registry Number | 135-16-0 |
|---|
| SMILES | NC1=NC(=O)C2=C(NC[C@H](CNC3=CC=C(C=C3)C(=O)NC(CCC(O)=O)C(O)=O)N2)N1 |
|---|
| InChI Identifier | InChI=1S/C19H23N7O6/c20-19-25-15-14(17(30)26-19)23-11(8-22-15)7-21-10-3-1-9(2-4-10)16(29)24-12(18(31)32)5-6-13(27)28/h1-4,11-12,21,23H,5-8H2,(H,24,29)(H,27,28)(H,31,32)(H4,20,22,25,26,30)/t11-,12?/m0/s1 |
|---|
| InChI Key | MSTNYGQPCMXVAQ-PXYINDEMSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Xanthines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthine
- Purinone
- 6-oxopurine
- Alkaloid or derivatives
- Pyrimidone
- Pyrimidine
- N-substituted imidazole
- Heteroaromatic compound
- Vinylogous amide
- Imidazole
- Azole
- Urea
- Lactam
- Azacycle
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Source: Biological location: |
|---|
| Role | Industrial application: |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 250 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 0.27 g/L | Not Available | | logP | -2.7 | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Tetrahydrofolic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0uka-4676900000-780e78301e85c240ab73 | Spectrum | | Predicted GC-MS | Tetrahydrofolic acid, 2 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-006x-1311190000-af98cc8be4da08f9b782 | Spectrum | | Predicted GC-MS | Tetrahydrofolic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0002-7796610000-0937514a7f23431b726e | 2018-05-25 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0nor-3269210000-20105ed6a7a6e24e2441 | 2018-05-25 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00kr-1935200000-a9449c19c1ae3b0d70da | 2018-05-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0032-0431900000-ff1f3948e47561161293 | 2016-06-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0952300000-ed9d84bc39d490f6210e | 2016-06-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001i-0920000000-0d0efcb6c323340d20b9 | 2016-06-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0001900000-abb4cec0eb79b0bd2de1 | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0v4l-2356900000-2fd17d2f587bbd5e882c | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9441000000-8c630c7a3633467d6bb3 | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0080900000-73480c24ce118e5e02f3 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-0491100000-0b5df4d0f168f8f15275 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0089-0962000000-9a988cf519f340f5d7cf | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-005c-0002900000-9587282a39f915386f95 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0zi3-2807900000-28355749389d1e225f7b | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0udi-3911000000-e7b1ccb0c9f2d7c8269a | 2021-09-24 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Pterine Biosynthesis |    | 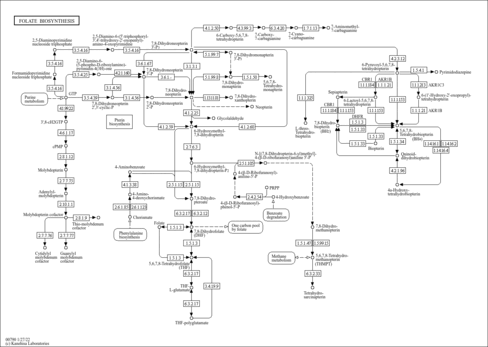 | | Folate Metabolism |    | 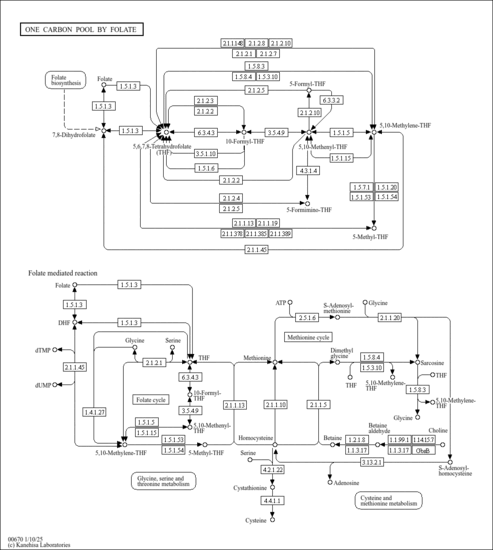 | | Glycine and Serine Metabolism | 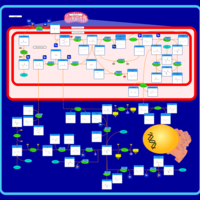 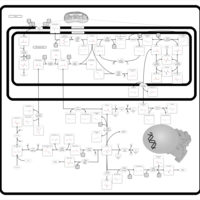 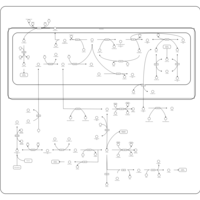 |  | | Betaine Metabolism |  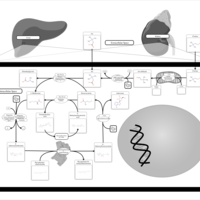  |  | | Histidine Metabolism | 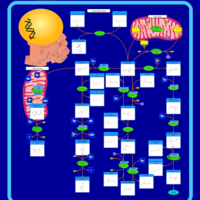 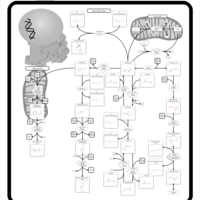 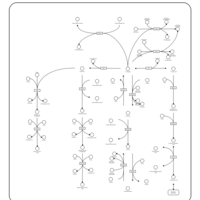 |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Methionine synthase | MTR | 1q43 | Q99707 | details | | Methylenetetrahydrofolate reductase | MTHFR | 1p36.3 | P42898 | details | | Trifunctional purine biosynthetic protein adenosine-3 | GART | 21q22.11 | P22102 | details | | Formimidoyltransferase-cyclodeaminase | FTCD | 21q22.3 | O95954 | details | | Aminomethyltransferase, mitochondrial | AMT | 3p21.2-p21.1 | P48728 | details | | Dihydrofolate reductase | DHFR | 5q11.2-q13.2 | P00374 | details | | Serine hydroxymethyltransferase, mitochondrial | SHMT2 | 12q12-q14 | P34897 | details | | Serine hydroxymethyltransferase, cytosolic | SHMT1 | 17p11.2 | P34896 | details | | Folylpolyglutamate synthase, mitochondrial | FPGS | 9q34.1 | Q05932 | details | | Bifunctional purine biosynthesis protein PURH | ATIC | 2q35 | P31939 | details | | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial | MTHFD2 | 2p13.1 | P13995 | details | | C-1-tetrahydrofolate synthase, cytoplasmic | MTHFD1 | 14q24 | P11586 | details | | Cytosolic 10-formyltetrahydrofolate dehydrogenase | ALDH1L1 | 3q21.3 | O75891 | details | | Glycine cleavage system H protein, mitochondrial | GCSH | | P23434 | details | | Gamma-glutamyl hydrolase | GGH | 8q12.3 | Q92820 | details | | Mitochondrial 10-formyltetrahydrofolate dehydrogenase | ALDH1L2 | 12q23.3 | Q3SY69 | details | | Methionyl-tRNA formyltransferase, mitochondrial | MTFMT | 15q22.31 | Q96DP5 | details | | Monofunctional C1-tetrahydrofolate synthase, mitochondrial | MTHFD1L | 6q25.1 | Q6UB35 | details | | Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase | MTHFD2 | 2p13.1 | Q7Z650 | details | | Dihydrofolate reductase, mitochondrial | DHFRL1 | 3q11.1 | Q86XF0 | details | | HemK methyltransferase family member 1 | HEMK1 | | Q9Y5R4 | details | | Methyltransferase-like protein 2B | METTL2B | | Q6P1Q9 | details | | Methyltransferase-like protein 6 | METTL6 | | Q8TCB7 | details | | Uncharacterized methyltransferase WBSCR22 | WBSCR22 | | O43709 | details |
|
|---|
| Transporters | |
| Cytosolic 10-formyltetrahydrofolate dehydrogenase | ALDH1L1 | 3q21.3 | O75891 | details | | Mitochondrial 10-formyltetrahydrofolate dehydrogenase | ALDH1L2 | 12q23.3 | Q3SY69 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0001846 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB022705 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 82572 |
|---|
| KEGG Compound ID | C00101 |
|---|
| BioCyc ID | THF |
|---|
| BiGG ID | 33856 |
|---|
| Wikipedia Link | Tetrahydrofolic acid |
|---|
| METLIN ID | 714 |
|---|
| PubChem Compound | 91443 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 20506 |
|---|
| References |
|---|
| General References | Not Available |
|---|



