| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:07:31 UTC |
|---|
| Updated at | 2020-11-18 16:39:18 UTC |
|---|
| CannabisDB ID | CDB005082 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Fructose 6-phosphate |
|---|
| Description | Fructose 6-phosphate, also known as neuberg ester or D-fructose-6-p, belongs to the class of organic compounds known as hexose phosphates. These are carbohydrate derivatives containing a hexose substituted by one or more phosphate groups. The open chain form of D-fructose 6-phosphate. Fructose 6-phosphate is an extremely weak basic (essentially neutral) compound (based on its pKa). Fructose 6-phosphate exists in all living species, ranging from bacteria to humans. fructose 6-phosphate and L-glutamine can be converted into glucosamine 6-phosphate and L-glutamic acid through the action of the enzyme glutamine--fructose-6-phosphate aminotransferase. In humans, fructose 6-phosphate is involved in homocarnosinosis. Outside of the human body, Fructose 6-phosphate has been detected, but not quantified in, carrots and milk (cow). This could make fructose 6-phosphate a potential biomarker for the consumption of these foods. Fructose 6-phosphate is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| D-Fructose 6-phosphate | ChEBI | | D-Fructose 6-phosphoric acid | ChEBI | | Fructose-6-phosphate | ChEBI | | Neuberg ester | ChEBI | | Fructose-6-phosphoric acid | Generator | | Fructose 6-phosphoric acid | Generator | | D-Fructose 6-phosphorate | HMDB | | D-Fructose-6-p | HMDB | | D-Fructose-6-phosphate | HMDB | | FPC | HMDB | | Fru-6-p | HMDB | | Fructose-6-p | HMDB | | Fructose-6P | HMDB | | Fructose-6-phosphate, (alpha-D)-isomer | HMDB | | Fructose-6-phosphate, (beta-D)-isomer | HMDB | | Fructose-6-phosphate, disodium salt, (D)-isomer | HMDB | | Fructose-6-phosphate, (D)-isomer | HMDB | | Fructose-6-phosphate, 1-(14)C-labeled, (D)-isomer | HMDB | | Fructose-6-phosphate, 2-(14)C-labeled, (D)-isomer | HMDB | | Fructose-6-phosphate, barium (1:1) salt, (D)-isomer | HMDB | | Fructose-6-phosphate, barium salt, (D)-isomer | HMDB | | Fructose-6-phosphate, sodium salt, (D)-isomer | HMDB | | Fructose-6-phosphate, calcium salt, (D)-isomer | HMDB | | Fructose-6-phosphate, magnesium (1:1) salt, (D)-isomer | HMDB | | Fructose-6-phosphate, sodium salt | HMDB |
|
|---|
| Chemical Formula | C6H13O9P |
|---|
| Average Molecular Weight | 260.14 |
|---|
| Monoisotopic Molecular Weight | 260.0297 |
|---|
| IUPAC Name | {[(2R,3R,4S)-2,3,4,6-tetrahydroxy-5-oxohexyl]oxy}phosphonic acid |
|---|
| Traditional Name | D-fructose 6-phosphate |
|---|
| CAS Registry Number | 643-13-0 |
|---|
| SMILES | OCC(=O)[C@@H](O)[C@H](O)[C@H](O)COP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H13O9P/c7-1-3(8)5(10)6(11)4(9)2-15-16(12,13)14/h4-7,9-11H,1-2H2,(H2,12,13,14)/t4-,5-,6-/m1/s1 |
|---|
| InChI Key | GSXOAOHZAIYLCY-HSUXUTPPSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as hexose phosphates. These are carbohydrate derivatives containing a hexose substituted by one or more phosphate groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Hexose phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hexose phosphate
- Monosaccharide phosphate
- Monoalkyl phosphate
- Acyloin
- Beta-hydroxy ketone
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Alpha-hydroxy ketone
- Ketone
- Secondary alcohol
- Polyol
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Primary alcohol
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Source: Biological location: |
|---|
| Role | Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 911 mg/mL | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | Fructose 6-phosphate, 6 TMS, GC-MS Spectrum | splash10-0gba-1943000000-9b9bb3f9964b519fa26b | Spectrum | | GC-MS | Fructose 6-phosphate, 6 TMS, GC-MS Spectrum | splash10-014j-1943000000-0be5d726668730e99be0 | Spectrum | | GC-MS | Fructose 6-phosphate, 6 TMS, GC-MS Spectrum | splash10-014j-1954000000-9fa83268db2925e9478d | Spectrum | | GC-MS | Fructose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-000b-0934000000-eb61ccae8d0b23564bc7 | Spectrum | | GC-MS | Fructose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-000b-0934000000-c2a49f4f2d11d3e60a9e | Spectrum | | GC-MS | Fructose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-000j-0925000000-a6b6a121cb447c890019 | Spectrum | | GC-MS | Fructose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-0f7a-1934000000-b28e738e5093324060d9 | Spectrum | | GC-MS | Fructose 6-phosphate, 1 MEOX; 6 TMS, GC-MS Spectrum | splash10-014i-3966000000-fee84a4ec7828d92e8b6 | Spectrum | | GC-MS | Fructose 6-phosphate, 1 MEOX; 6 TMS, GC-MS Spectrum | splash10-014i-1955000000-32da70382eccbcbbb574 | Spectrum | | GC-MS | Fructose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-0gba-1943000000-9b9bb3f9964b519fa26b | Spectrum | | GC-MS | Fructose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-014j-1943000000-0be5d726668730e99be0 | Spectrum | | GC-MS | Fructose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-014j-1954000000-9fa83268db2925e9478d | Spectrum | | GC-MS | Fructose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-000b-0934000000-eb61ccae8d0b23564bc7 | Spectrum | | GC-MS | Fructose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-000b-0934000000-c2a49f4f2d11d3e60a9e | Spectrum | | GC-MS | Fructose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-000j-0925000000-a6b6a121cb447c890019 | Spectrum | | GC-MS | Fructose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-0f7a-1934000000-b28e738e5093324060d9 | Spectrum | | GC-MS | Fructose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-014i-3966000000-fee84a4ec7828d92e8b6 | Spectrum | | GC-MS | Fructose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-014i-1955000000-32da70382eccbcbbb574 | Spectrum | | GC-MS | Fructose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-0gba-1933000000-1c8febd80da1382bec31 | Spectrum | | Predicted GC-MS | Fructose 6-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0k97-9840000000-4f9f16f21249d9870dfa | Spectrum | | Predicted GC-MS | Fructose 6-phosphate, 4 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00o9-7934540000-e23e661c1babf5e063a8 | Spectrum | | Predicted GC-MS | Fructose 6-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Fructose 6-phosphate, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Fructose 6-phosphate, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Fructose 6-phosphate, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0229-5090000000-281699f4bce06db0b1b0 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-006x-9730000000-b72576e6bb1ae73e6881 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0kbf-9740000000-130bb715a91e870b5480 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0aor-0690000000-b640c7fba12f06d6bfa2 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-00kb-9710000000-5d6429381cefb6a2bdc7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0002-9100000000-51c3f5ca7ef3f190f1df | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-054k-9000000000-75c50b162633f723546d | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-004i-9000000000-07895e6c6f3e4a816391 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0aor-0690000000-b640c7fba12f06d6bfa2 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0002-9000000000-f31b175fc6a9941a2451 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9220000000-962294aed3032864dc90 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-c2dc78595233b4d4df78 | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dl-3590000000-3fc4dac2f46082421870 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0c04-9610000000-77e7a6ced5ab4dda6f86 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9200000000-e4500e644878a4632447 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0ar9-9720000000-7ca18e5a32ef069ff21a | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9200000000-00032f32c05509571a9f | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-5e859d2084880df68725 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000b-9620000000-1a2150ea2d68dcce9b24 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-002b-9000000000-f2969657deda9ac99077 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-d5aaca13a836cb9f1725 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dm-2980000000-ccecfa1c0258f9267846 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9100000000-550809f9719b91a7807a | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9000000000-501178ab2f5d5affab48 | 2021-09-24 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Glycolysis | 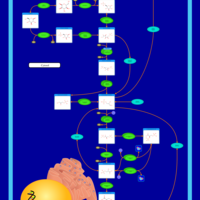 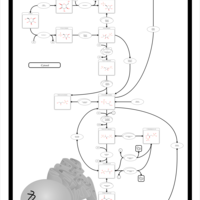 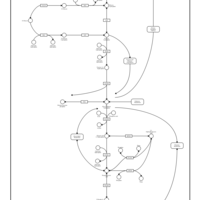 |  | | Gluconeogenesis | 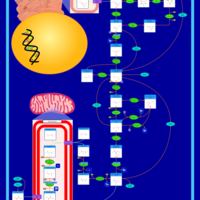 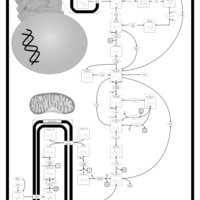 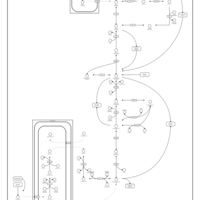 |  | | Fructose and Mannose Degradation | 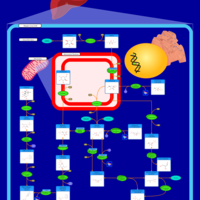 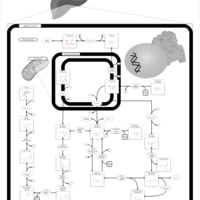 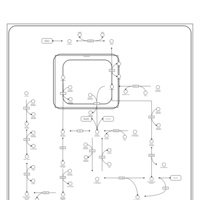 |  | | Pentose Phosphate Pathway | 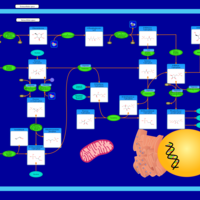  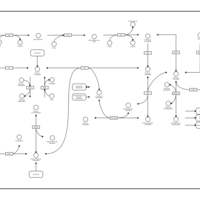 |  | | Amino Sugar Metabolism | 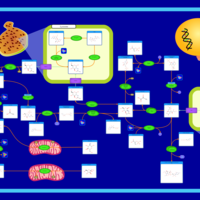 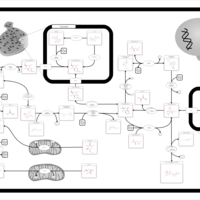 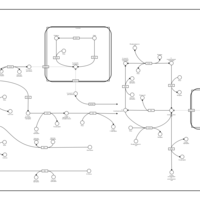 |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000124 |
|---|
| DrugBank ID | DB04493 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB021896 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 62713 |
|---|
| KEGG Compound ID | C00085 |
|---|
| BioCyc ID | CPD-15709 |
|---|
| BiGG ID | 33795 |
|---|
| Wikipedia Link | Fructose 6-phosphate |
|---|
| METLIN ID | 5159 |
|---|
| PubChem Compound | 69507 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15946 |
|---|
| References |
|---|
| General References | Not Available |
|---|



