| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:07:19 UTC |
|---|
| Updated at | 2020-11-18 16:39:17 UTC |
|---|
| CannabisDB ID | CDB005080 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Glucose 6-phosphate |
|---|
| Description | Glucose 6-phosphate, also known as robison ester or GLC6P, belongs to the class of organic compounds known as hexose phosphates. These are carbohydrate derivatives containing a hexose substituted by one or more phosphate groups. Glucose 6-phosphate is an extremely weak basic (essentially neutral) compound (based on its pKa). Glucose 6-phosphate exists in all living species, ranging from bacteria to humans. Within humans, glucose 6-phosphate participates in a number of enzymatic reactions. In particular, glucose 6-phosphate can be converted into D-glucose through the action of the enzyme glucose-6-phosphatase. In addition, glucose 6-phosphate can be biosynthesized from glucose 1-phosphate through its interaction with the enzyme phosphoglucomutase-1. In humans, glucose 6-phosphate is involved in the metabolic disorder called the glycogen storage disease type 1A (gsd1a) or von gierke disease pathway. A glucopyranose ring with a phosphate replacing the hydroxy in the hydroxymethyl group at position 6. Glucose 6-phosphate is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 6-O-Phosphono-D-glucopyranose | ChEBI | | D-Glucose 6-phosphate | ChEBI | | GLC6p | ChEBI | | Robison ester | ChEBI | | D-Glucose 6-phosphoric acid | Generator | | Glucose 6-phosphoric acid | Generator | | a-D-Glucose 6- phosphate | HMDB | | alpha-D-Glucose 6- phosphate | HMDB | | alpha-D-Glucose 6-phosphate | HMDB | | alpha-D-Hexose 6-phosphate | HMDB | | D(+)-Glucopyranose 6-phosphate | HMDB | | D-Glucose-6-dihydrogen phosphate | HMDB | | D-Hexose 6-phosphate | HMDB | | Glucose-6-phosphate | HMDB | | Glucose 6-phosphate | KEGG | | D-Glucopyranose 6-phosphoric acid | Generator |
|
|---|
| Chemical Formula | C6H13O9P |
|---|
| Average Molecular Weight | 260.14 |
|---|
| Monoisotopic Molecular Weight | 260.0297 |
|---|
| IUPAC Name | {[(2R,3S,4S,5R)-3,4,5,6-tetrahydroxyoxan-2-yl]methoxy}phosphonic acid |
|---|
| Traditional Name | glucose 6-phosphate |
|---|
| CAS Registry Number | 56-73-5 |
|---|
| SMILES | OC1O[C@H](COP(O)(O)=O)[C@@H](O)[C@H](O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C6H13O9P/c7-3-2(1-14-16(11,12)13)15-6(10)5(9)4(3)8/h2-10H,1H2,(H2,11,12,13)/t2-,3-,4+,5-,6?/m1/s1 |
|---|
| InChI Key | NBSCHQHZLSJFNQ-GASJEMHNSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as hexose phosphates. These are carbohydrate derivatives containing a hexose substituted by one or more phosphate groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Hexose phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hexose phosphate
- Monosaccharide phosphate
- Monoalkyl phosphate
- Organic phosphoric acid derivative
- Alkyl phosphate
- Oxane
- Phosphoric acid ester
- Hemiacetal
- Secondary alcohol
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Hydrocarbon derivative
- Alcohol
- Organic oxide
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Source: Biological location: |
|---|
| Physical Properties |
|---|
| State | Liquid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | Glucose 6-phosphate, 6 TMS, GC-MS Spectrum | splash10-000b-1932000000-59f4881b86595037d013 | Spectrum | | GC-MS | Glucose 6-phosphate, 6 TMS, GC-MS Spectrum | splash10-0f7t-1933000000-4894135f21c21c369428 | Spectrum | | GC-MS | Glucose 6-phosphate, 6 TMS; 1 MEOX, GC-MS Spectrum | splash10-00di-9522000000-81266970683faa1f2aaf | Spectrum | | GC-MS | Glucose 6-phosphate, 6 TMS; 1 MEOX, GC-MS Spectrum | splash10-00di-9412000000-2f2a0e772025b48bf6f3 | Spectrum | | GC-MS | Glucose 6-phosphate, 1 MEOX; 6 TMS, GC-MS Spectrum | splash10-000j-1956000000-692b4df2b9c16f6a780c | Spectrum | | GC-MS | Glucose 6-phosphate, 1 MEOX; 6 TMS, GC-MS Spectrum | splash10-000i-1957000000-bd94955ffc1474153a48 | Spectrum | | GC-MS | Glucose 6-phosphate, 6 TMS, GC-MS Spectrum | splash10-0v0a-0596000000-676527ddfc1d596e78f8 | Spectrum | | GC-MS | Glucose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-000b-1932000000-59f4881b86595037d013 | Spectrum | | GC-MS | Glucose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-0f7t-1933000000-4894135f21c21c369428 | Spectrum | | GC-MS | Glucose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-00di-9522000000-81266970683faa1f2aaf | Spectrum | | GC-MS | Glucose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-00di-9412000000-2f2a0e772025b48bf6f3 | Spectrum | | GC-MS | Glucose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-000j-1956000000-692b4df2b9c16f6a780c | Spectrum | | GC-MS | Glucose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-000i-1957000000-bd94955ffc1474153a48 | Spectrum | | GC-MS | Glucose 6-phosphate, non-derivatized, GC-MS Spectrum | splash10-0v0a-0596000000-676527ddfc1d596e78f8 | Spectrum | | Predicted GC-MS | Glucose 6-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0005-9740000000-a6e81224fb97751dc2e6 | Spectrum | | Predicted GC-MS | Glucose 6-phosphate, 4 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-001i-6252790000-ef5662fb1886367e07f2 | Spectrum | | Predicted GC-MS | Glucose 6-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Glucose 6-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, N/A (Annotated) | splash10-03di-0090000000-5653def80d1b5ff0eebb | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, N/A (Annotated) | splash10-000b-9200000000-66741d773ac31b347625 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, N/A (Annotated) | splash10-000t-9000000000-068809fcfaff0d387e6b | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0a4i-1090000000-d6e539d78b38c01dafea | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0002-9210000000-8b9ff1c5095f48a81fef | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0002-9000000000-f6e1147852f75db4c0a4 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-002b-9000000000-1c52fd659f8a2b795e51 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-004i-9000000000-d3f6a05e18ea6d56c56d | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0006-0890000000-5f3047c92c970dfe6927 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-004i-0900000000-b84eb7685667fa7ee85e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-004i-2900000000-d4a0d51bb6e2da28533a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-004i-6900000000-dfb5bcbd9a7439c32352 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-03gi-9600000000-a0922c82e185f6b5aa67 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-0006-0290000000-7ac7b397c52f29cd3822 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-052b-6590000000-5cdbc23378480ac55c4f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-004i-1900000000-b74270f21c6086362227 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-002b-9000000000-bc69e8fa215a3b82dcf2 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0a4i-0090000000-d6e539d78b38c01dafea | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0002-9210000000-10019f92fd1eca143bab | 2017-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0490000000-94ea79ec673f4688b7a0 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dm-5940000000-c649262446626dcb8d6e | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0uka-9800000000-aae2cb792906d2dec6cd | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a6r-8290000000-8e20219f94b3fe964e38 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-b19b64f5cb4124047baf | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-ecb75a44e3d25affdf31 | 2016-09-12 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Gluconeogenesis | 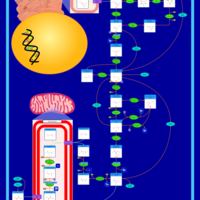 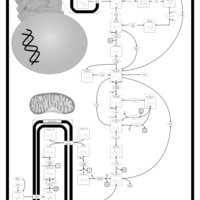 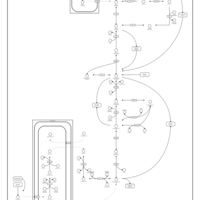 |  | | Pentose Phosphate Pathway | 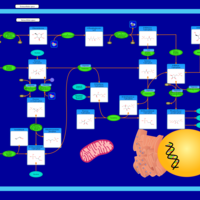  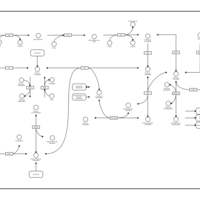 |  | | Inositol Metabolism | 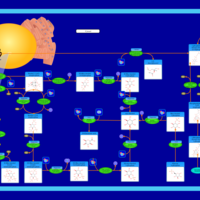 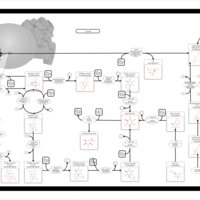 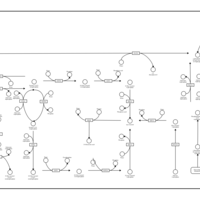 |  | | Starch and Sucrose Metabolism | 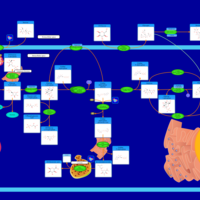 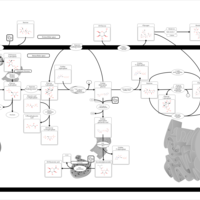 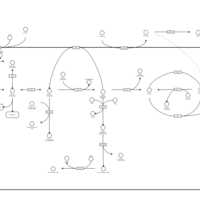 |  | | Glycolysis | 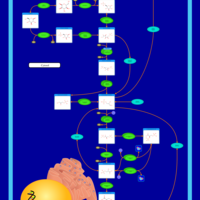 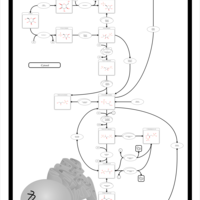 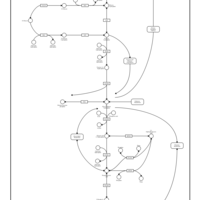 |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0001401 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB021818 |
|---|
| KNApSAcK ID | C00007306 |
|---|
| Chemspider ID | 5743 |
|---|
| KEGG Compound ID | C00092 |
|---|
| BioCyc ID | D-glucose-6-phosphate |
|---|
| BiGG ID | 36977 |
|---|
| Wikipedia Link | Glucose_6-phosphate |
|---|
| METLIN ID | 145 |
|---|
| PubChem Compound | 5958 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 4170 |
|---|
| References |
|---|
| General References | Not Available |
|---|



