| Identification |
|---|
| HMDB Protein ID
| CDBP00362 |
| Secondary Accession Numbers
| Not Available |
| Name
| Glycogen phosphorylase, liver form |
| Description
| Not Available |
| Synonyms
|
Not Available
|
| Gene Name
| PYGL |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in phosphorylase activity |
| Specific Function
| Phosphorylase is an important allosteric enzyme in carbohydrate metabolism. Enzymes from different sources differ in their regulatory mechanisms and in their natural substrates. However, all known phosphorylases share catalytic and structural properties.
|
| GO Classification
|
| Biological Process |
| 5-phosphoribose 1-diphosphate biosynthetic process |
| glucose metabolic process |
| glucose homeostasis |
| glycogen catabolic process |
| carbohydrate metabolic process |
| small molecule metabolic process |
| Cellular Component |
| cytosol |
| cytoplasm |
| plasma membrane |
| Function |
| binding |
| catalytic activity |
| transferase activity |
| cofactor binding |
| pyridoxal phosphate binding |
| transferase activity, transferring hexosyl groups |
| phosphorylase activity |
| transferase activity, transferring glycosyl groups |
| Molecular Function |
| glucose binding |
| ATP binding |
| pyridoxal phosphate binding |
| bile acid binding |
| AMP binding |
| protein homodimerization activity |
| purine nucleobase binding |
| drug binding |
| glycogen phosphorylase activity |
| vitamin binding |
| Process |
| metabolic process |
| primary metabolic process |
| carbohydrate metabolic process |
|
| Cellular Location
|
Not Available
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Insulin signaling pathway | Not Available | 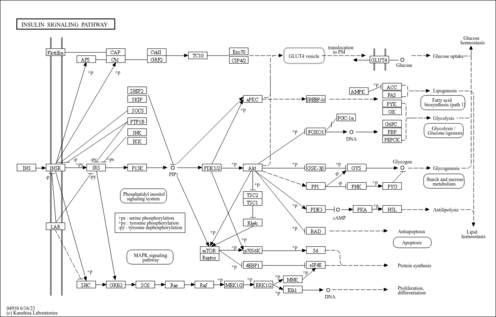 | | Starch and Sucrose Metabolism |    |  | | Glycogen synthetase deficiency | 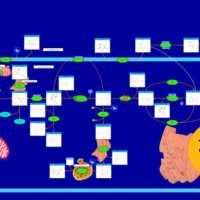   | Not Available | | Glycogenosis, Type III. Cori disease, Debrancher glycogenosis | 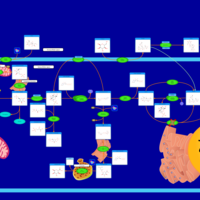   | Not Available | | Glycogenosis, Type IV. Amylopectinosis, Anderson disease |    | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 14 |
| Locus
| 14q21-q22 |
| SNPs
| PYGL |
| Gene Sequence
|
>2544 bp
ATGGGCGAACCGCTGACAGACCAGGAGAAGCGGCGGCAGATCAGCATCCGCGGCATCGTG
GGCGTGGAGAACGTGGCAGAGCTGAAGAAGAGTTTCAACCGGCACCTGCACTTCACGCTG
GTCAAGGACCGCAACGTGGCCACCACCCGCGACTACTACTTCGCGCTGGCGCACACGGTG
CGGGACCACCTGGTGGGGCGCTGGATCCGCACGCAGCAGCACTACTACGACAAGTGCCCC
AAGAGGGAATATTACCTCTCTCTGGAATTTTACATGGGCCGAACATTACAGAACACCATG
ATCAACCTCGGTCTGCAAAATGCCTGTGATGAGGCCATTTACCAGCTTGGATTGGATATA
GAAGAGTTAGAAGAAATTGAAGAAGATGCTGGACTTGGCAATGGTGGTCTTGGGAGACTT
GCTGCCTGCTTCTTGGATTCCATGGCAACCCTGGGACTTGCAGCCTATGGATACGGCATT
CGGTATGAATATGGGATTTTCAATCAGAAGATCCGAGATGGATGGCAGGTAGAAGAAGCA
GATGATTGGCTCAGATATGGAAACCCTTGGGAGAAGTCCCGCCCAGAATTCATGCTGCCT
GTGCACTTCTATGGAAAAGTAGAACACACCAACACCGGGACCAAGTGGATTGACACTCAA
GTGGTCCTGGCTCTGCCATATGACACCCCCGAGCCCGGCTACATGAATAACACTGTCAAC
ACCATGCGCCTCTGGTCTGCTCGGGCACCAAATGACTTTAACCTCAGAGACTTTAATGTT
GGAGACTACATTCAGGCTGTGCTGGACCGAAACCTGGCCGAGAACATCTCCCGGGTCCTC
TATCCCAATGACAATTTTTTTGAAGGGAAGGAGCTAAGATTGAAGCAGGAATACTTTGTG
GTGGCTGCAACCTTGCAAGATATCATCCGCCGTTTCAAAGCCTCCAAGTTTGGCTCCACC
CGTGGTCAAGGAACTGTGTTTGATGCCTTCCCGGATCAGGTGGCCATCCAGCTGAATGAT
ACTCACCCTCGCATCGCGATCCCTGAGCTGATGAGGATTTTTGTGGATATTGAAAAACTG
CCCTGGTCCAAGGCATGGGAGCTCAACCAGAAGACCTTCGCCTACACCAACCACACAGTG
CTCCCGGAAGCCCTGGAGCGCTGGCCCGTGGACCTGGTGGAGAAGCTGCTCCCTCGACAT
TTGGAAATCATTTATGAGATAAATCAGAAGCATTTAGATAGAATTGTGGCCTTGTTTCCT
AAAGATGTGGACCCTCTGAGAAGGATGTCTCTGATAGAAGAGGAAGGAAGCAAAAGGATC
AACATGGCCCATCTCTGCATTGTCGGTTCCCATGCTGTGAATGGCGTGGCTAAAATCCAC
TCAGACATCGTGAAGACTAAAGTATTCAAGGACTTCAGTGAGCTAGAACCTGACAAGTTT
CAGAATAAAACCAATGGGATCACTCCAAGGCGCTGGCTCCTACTCTGCAACCCAGGACTT
GCAGAGCTCATAGCAGAGAAAATTGGAGAAGACTATGTGAAAGACCTGAGCCAGCTGACG
AAGCTCCACAGCTTCCTGGGTGATGATGTCTTCCTCCGGGAACTCGCCAAGGTGAAGCAG
GAGAATAAGCTGAAGTTTTCTCAGTTCCTGGAGACGGAGTACAAAGTGAAGATCAACCCA
TCCTCCATGTTTGATGTCCAGGTGAAGAGGATACATGAGTACAAGCGACAGCTCTTGAAC
TGTCTGCATGTGATCACGATGTACAACCGCATTAAGAAAGACCCTAAGAAGTTATTCGTG
CCAAGGACAGTTATCATTGGTGGTAAAGCTGCCCCAGGATATCACATGGCCAAAATGATC
ATAAAGCTGATCACTTCAGTGGCAGATGTGGTGAACAATGACCCTATGGTTGGAAGCAAG
TTGAAAGTCATCTTCTTGGAGAACTACAGAGTATCTCTTGCTGAAAAAGTCATTCCAGCC
ACAGATCTGTCAGAGCAGATTTCCACTGCAGGCACCGAAGCCTCGGGGACAGGCAATATG
AAGTTCATGCTAAATGGGGCCCTAACTATCGGGACCATGGATGGGGCCAATGTGGAAATG
GCAGAAGAAGCTGGGGAAGAGAACCTGTTCATCTTTGGCATGAGCATAGATGATGTGGCT
GCTTTGGACAAGAAAGGGTACGAGGCAAAAGAATACTATGAGGCACTTCCAGAGCTGAAG
CTGGTCATTGATCAAATTGACAATGGCTTTTTTTCTCCCAAGCAGCCTGACCTCTTCAAA
GATATCATCAACATGCTATTTTATCATGACAGGTTTAAAGTCTTTGCAGACTACGAAGCC
TATGTCAAGTGTCAAGATAAAGTGAGTCAGCTGTACATGAATCCAAAGGCCTGGAACACA
ATGGTACTCAAAAACATAGCTGCCTCGGGGAAATTCTCCAGTGACCGAACAATTAAAGAA
TATGCCCAAAACATCTGGAACGTGGAACCTTCAGATCTAAAGATTTCTCTATCCAATGAA
TCTAACAAAGTCAATGGAAATTGA
|
| Protein Properties |
|---|
| Number of Residues
| 847 |
| Molecular Weight
| 93133.25 |
| Theoretical pI
| 7.303 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Glycogen phosphorylase, liver form
MAKPLTDQEKRRQISIRGIVGVENVAELKKSFNRHLHFTLVKDRNVATTRDYYFALAHTV
RDHLVGRWIRTQQHYYDKCPKRVYYLSLEFYMGRTLQNTMINLGLQNACDEAIYQLGLDI
EELEEIEEDAGLGNGGLGRLAACFLDSMATLGLAAYGYGIRYEYGIFNQKIRDGWQVEEA
DDWLRYGNPWEKSRPEFMLPVHFYGKVEHTNTGTKWIDTQVVLALPYDTPVPGYMNNTVN
TMRLWSARAPNDFNLRDFNVGDYIQAVLDRNLAENISRVLYPNDNFFEGKELRLKQEYFV
VAATLQDIIRRFKASKFGSTRGAGTVFDAFPDQVAIQLNDTHPALAIPELMRIFVDIEKL
PWSKAWELTQKTFAYTNHTVLPEALERWPVDLVEKLLPRHLEIIYEINQKHLDRIVALFP
KDVDRLRRMSLIEEEGSKRINMAHLCIVGSHAVNGVAKIHSDIVKTKVFKDFSELEPDKF
QNKTNGITPRRWLLLCNPGLAELIAEKIGEDYVKDLSQLTKLHSFLGDDVFLRELAKVKQ
ENKLKFSQFLETEYKVKINPSSMFDVQVKRIHEYKRQLLNCLHVITMYNRIKKDPKKLFV
PRTVIIGGKAAPGYHMAKMIIKLITSVADVVNNDPMVGSKLKVIFLENYRVSLAEKVIPA
TDLSEQISTAGTEASGTGNMKFMLNGALTIGTMDGANVEMAEEAGEENLFIFGMRIDDVA
ALDKKGYEAKEYYEALPELKLVIDQIDNGFFSPKQPDLFKDIINMLFYHDRFKVFADYEA
YVKCQDKVSQLYMNPKAWNTMVLKNIAASGKFSSDRTIKEYAQNIWNVEPSDLKISLSNE
SNKVNGN
|
| External Links |
|---|
| GenBank ID Protein
| 183353 |
| UniProtKB/Swiss-Prot ID
| P06737 |
| UniProtKB/Swiss-Prot Entry Name
| PYGL_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| M14636 |
| GeneCard ID
| PYGL |
| GenAtlas ID
| PYGL |
| HGNC ID
| HGNC:9725 |
| References |
|---|
| General References
| Not Available |

