| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:51:39 UTC |
|---|
| Updated at | 2022-12-13 23:36:28 UTC |
|---|
| CannabisDB ID | CDB004930 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Salicylic acid |
|---|
| Description | Salicylic acid, also known as 2-carboxyphenol or 2-hydroxybenzoate, belongs to the class of organic compounds known as salicylic acids. These are ortho-hydroxylated benzoic acids. Salicylic acid is also classified as a beta hydroxy acid (BHA). Salicylic acid is a weak acid and exists as a white powder and has no detectable odor. It is readily soluble in water. Salicylic acid was originally isolated from willow trees and is found in many other plants where it functions as a plant hormone or phytohormone to facilitate plant defenses against pathogens (PMID: 31489905 ). Salicylic acid also helps plants cope with other abiotic stressors such a chilling, freezing, heat, heavy metals, salt and drought (PMID: 31489905 ). Salicylic acid also plays a role in the regulation or stomatal closure and seed germination. Industrially, salicylic acid is used as a food preservative, a bactericidal and an antiseptic. It is also used in the production of various pharmaceuticals, including 4-aminosalicylic acid, sandulpiride, landetimide and most notably acetylsalicylic acid or ASA (aspirin). Medically, salicylic acid is used most commonly to help remove the outer layer of the skin. As such, it is used to treat warts, psoriasis, acne, ringworm, dandruff, and ichthyosis. Salicylic acid modulates COX2 gene expression to decrease the formation of pro-inflammatory prostaglandins. Salicylic acid also inhibits the oxidation of uridine-5-diphosphoglucose (UDPG) competitively with nicotinamide adenosine dinucleotide (NAD). In humans, salicylic acid is involved in salicylic acid action pathway. Salicylic acid has been detected, but not quantified in, several different foods and plants, such as canola, cocoa beans, rowanberries, yautia, and ucuhuba. This could make salicylic acid a potential biomarker for the consumption of these foods. Salicylic acid is also found in cannabis smoke and is formed during the combustion of cannabis ( Ref:DOI ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Carboxyphenol | ChEBI | | 2-HYDROXYBENZOIC ACID | ChEBI | | O-Carboxyphenol | ChEBI | | O-Hydroxybenzoic acid | ChEBI | | 2-HYDROXYBENZOate | Generator | | O-Hydroxybenzoate | Generator | | Salicylate | Generator | | 2 Hydroxybenzoic acid | MeSH | | Acid, 2-hydroxybenzoic | MeSH | | Acid, salicylic | MeSH | | Acid, O-hydroxybenzoic | MeSH | | Acid, ortho-hydroxybenzoic | MeSH | | O Hydroxybenzoic acid | MeSH | | Ortho hydroxybenzoic acid | MeSH | | Ortho-hydroxybenzoic acid | MeSH | | 2-Hydroxybenzenecarboxylate | HMDB | | 2-Hydroxybenzenecarboxylic acid | HMDB | | Advanced pain relief callus removers | HMDB | | Advanced pain relief corn removers | HMDB | | Clear away wart remover | HMDB | | Compound W | HMDB | | Dr. scholl's callus removers | HMDB | | Dr. scholl's corn removers | HMDB | | Dr. scholl's wart remover kit | HMDB | | Duofil wart remover | HMDB | | Duoplant | HMDB | | Freezone | HMDB | | Ionil | HMDB | | Ionil plus | HMDB | | K 537 | HMDB | | K 557 | HMDB | | Phenol-2-carboxylate | HMDB | | Phenol-2-carboxylic acid | HMDB | | Psoriacid-S-stift | HMDB | | Retarder W | HMDB | | Rutranex | HMDB | | Salicylic acid collodion | HMDB | | Salicylic acid soap | HMDB | | Saligel | HMDB | | Salonil | HMDB | | Stri-dex | HMDB | | trans-Ver-sal | HMDB | | SA | PhytoBank |
|
|---|
| Chemical Formula | C7H6O3 |
|---|
| Average Molecular Weight | 138.12 |
|---|
| Monoisotopic Molecular Weight | 138.0317 |
|---|
| IUPAC Name | 2-hydroxybenzoic acid |
|---|
| Traditional Name | salicylic |
|---|
| CAS Registry Number | 69-72-7 |
|---|
| SMILES | OC(=O)C1=CC=CC=C1O |
|---|
| InChI Identifier | InChI=1S/C7H6O3/c8-6-4-2-1-3-5(6)7(9)10/h1-4,8H,(H,9,10) |
|---|
| InChI Key | YGSDEFSMJLZEOE-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as salicylic acids. These are ortho-hydroxylated benzoic acids. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Benzenoids |
|---|
| Class | Benzene and substituted derivatives |
|---|
| Sub Class | Benzoic acids and derivatives |
|---|
| Direct Parent | Salicylic acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Salicylic acid
- Benzoic acid
- Benzoyl
- 1-hydroxy-4-unsubstituted benzenoid
- 1-hydroxy-2-unsubstituted benzenoid
- Phenol
- Vinylogous acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 158 °C | Not Available | | Boiling Point | 200 °C | Wikipedia | | Water Solubility | 2.24 mg/mL at 25 °C | Not Available | | logP | 2.26 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-00du-9600000000-6d4a0ff2d48d814b5c54 | 2014-09-20 | View Spectrum | | GC-MS | Salicylic acid, 2 TMS, GC-MS Spectrum | splash10-014i-3890000000-62eae168a9d7ab3ada6f | Spectrum | | GC-MS | Salicylic acid, non-derivatized, GC-MS Spectrum | splash10-00du-9700000000-e1e2ee6b61d86c596403 | Spectrum | | GC-MS | Salicylic acid, non-derivatized, GC-MS Spectrum | splash10-014i-3890000000-62eae168a9d7ab3ada6f | Spectrum | | GC-MS | Salicylic acid, non-derivatized, GC-MS Spectrum | splash10-014i-2960000000-1b6b46cbb2b643b71448 | Spectrum | | Predicted GC-MS | Salicylic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0079-8900000000-e8ee46d81fcc1ce3766e | Spectrum | | Predicted GC-MS | Salicylic acid, 2 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-006x-8950000000-9ed3a56f2b2654ba281f | Spectrum | | Predicted GC-MS | Salicylic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Salicylic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-059j-9600000000-54545731fceee84be340 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-00xu-9500000000-2f1c989b672669aaf083 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-0gb9-9000000000-a0049e982e8ecd7ab730 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-000i-0900000000-f1e71df6894bcc8dda74 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0006-9200000000-f9fd317c182ec7ca90dc | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0006-9000000000-2b17aea4ee0ddd6321cf | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0006-9000000000-320b7cd879b61439cf42 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0006-9000000000-7d1b96d60026076a7ecc | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - ESI-TOF , Negative | splash10-002b-0496100000-97708001d2a6d031beff | 2017-08-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - ESI-TOF 10V, Negative | splash10-002b-0496100000-97708001d2a6d031beff | 2017-08-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - ESI-TOF , Negative | splash10-002b-0496100000-97708001d2a6d031beff | 2017-09-12 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - ESI-TOF 10V, Negative | splash10-000i-0900000000-f88c693bac9b89416a52 | 2017-09-12 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-0006-9400000000-b0fb5458dfa73429b976 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-0006-9400000000-b0fb5458dfa73429b976 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-0006-9100000000-237ee14e8af5262c0dab | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-0006-9000000000-3ec5d7a9114e37b8af2a | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-0006-9000000000-d8fdab29114453b10280 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-0006-9000000000-4a337e3639c9f42a9000 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-0006-9000000000-2deb0e843e099fdcea99 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-00kf-9000000000-951b374b351c5534628a | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-014l-9000000000-4ea52d5aa4344a40c007 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-000i-0900000000-f1e71df6894bcc8dda74 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0006-9200000000-65f3f297188dc07b53be | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0006-9000000000-2b17aea4ee0ddd6321cf | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0006-9000000000-320b7cd879b61439cf42 | 2017-09-14 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Salicylic Acid Action Pathway | 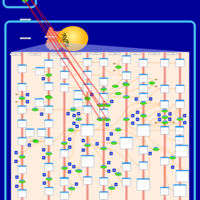 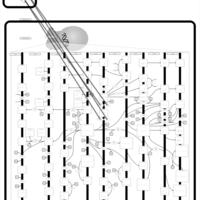 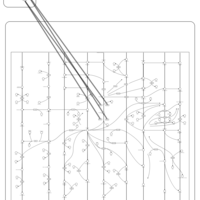 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 0.0231 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 0.0236 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 0.0103 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.0214 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 0.0104 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 0.00854 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0001895 |
|---|
| DrugBank ID | DB00936 |
|---|
| Phenol Explorer Compound ID | 428 |
|---|
| FoodDB ID | FDB000882 |
|---|
| KNApSAcK ID | C00000206 |
|---|
| Chemspider ID | 331 |
|---|
| KEGG Compound ID | C00805 |
|---|
| BioCyc ID | CPD-110 |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Salicylic_Acid |
|---|
| METLIN ID | 616 |
|---|
| PubChem Compound | 338 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16914 |
|---|
| References |
|---|
| General References | - Pokotylo I, Kravets V, Ruelland E: Salicylic Acid Binding Proteins (SABPs): The Hidden Forefront of Salicylic Acid Signalling. Int J Mol Sci. 2019 Sep 6;20(18). pii: ijms20184377. doi: 10.3390/ijms20184377. [PubMed:31489905 ]
|
|---|