| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-19 00:26:05 UTC |
|---|
| Updated at | 2022-12-13 23:36:24 UTC |
|---|
| CannabisDB ID | CDB000235 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Aspartic acid |
|---|
| Description | Aspartic acid (Asp, D), also known as aspartate, the name of its anion, is one of the 20 natural proteinogenic amino acids which are the building blocks of proteins. As such, it is also present in Cannabis sativa (PMID: 6991645 ). As its name indicates, aspartic acid is the carboxylic acid analog of asparagine. As a neurotransmitter, aspartic acid may provide resistance to fatigue and thus lead to endurance, although the evidence to support this idea is not strong. Aspartic acid is a nonessential amino acid, since it can be made from glutamic acid by enzymes using vitamin B6. The amino acid has important roles in the urea cycle and DNA metabolism. Aspartic acid is a major excitatory neurotransmitter, which is sometimes found to be increased in epileptic and stroke patients. It is decreased in depressed patients and in patients with brain atrophy. Aspartic acid supplements are being evaluated. Five grams can raise blood levels. Magnesium and zinc may be natural inhibitors of some of the actions of aspartic acid. Aspartic acid, with the amino acid phenylalanine, is a part of the natural sweetener, aspartame. This sweetener is an advance in artificial sweeteners and is probably safe in normal doses to all except phenylketonurics. The jury is still out on the long-term effects it has on many brain neurohormones. Aspartic acid may be a significant immunostimulant of the thymus and can protect against some of the damaging effects of radiation. Many claims have been made for the special value of administering aspartic acid in the form of potassium and magnesium salts. Since aspartic acid is relatively nontoxic, studies are now in progress to elucidate its pharmacological and therapeutic roles. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (S)-2-Aminobutanedioic acid | ChEBI | | (S)-2-Aminosuccinic acid | ChEBI | | 2-Aminosuccinic acid | ChEBI | | Asp | ChEBI | | ASPARTIC ACID | ChEBI | | D | ChEBI | | L-Asparaginsaeure | ChEBI | | L-Asp | Kegg | | (S)-2-Aminobutanedioate | Generator | | (S)-2-Aminosuccinate | Generator | | 2-Aminosuccinate | Generator | | ASPARTate | Generator | | L-Aspartate | Generator | | (+)-Aspartate | HMDB | | (+)-Aspartic acid | HMDB | | (2S)-Aspartate | HMDB | | (2S)-Aspartic acid | HMDB | | (L)-Aspartate | HMDB | | (L)-Aspartic acid | HMDB | | (R)-2-Aminosuccinate | HMDB | | (S)-(+)-Aspartate | HMDB | | (S)-(+)-Aspartic acid | HMDB | | (S)-Amino-butanedioate | HMDB | | (S)-Amino-butanedioic acid | HMDB | | (S)-Aminobutanedioate | HMDB | | (S)-Aminobutanedioic acid | HMDB | | (S)-Aspartate | HMDB | | (S)-Aspartic acid | HMDB | | 2-Amino-3-methylsuccinate | HMDB | | 2-Amino-3-methylsuccinic acid | HMDB | | alpha-Aminosuccinate | HMDB | | alpha-Aminosuccinic acid | HMDB | | Aminosuccinate | HMDB | | Asparagate | HMDB | | Asparagic acid | HMDB | | Asparaginate | HMDB | | Asparaginic acid | HMDB | | Asparatate | HMDB | | H-Asp-OH | HMDB | | L-(+)-Aspartate | HMDB | | L-(+)-Aspartic acid | HMDB | | L-Aminosuccinate | HMDB | | L-Aminosuccinic acid | HMDB | | L-Asparagate | HMDB | | L-Asparagic acid | HMDB | | L-Asparaginate | HMDB | | L-Asparaginic acid | HMDB | | (+-)-Aspartic acid | HMDB | | (R,S)-Aspartic acid | HMDB | | Aspartate, disodium | HMDB | | Aspartate, magnesium | HMDB | | Aspartate, monopotassium | HMDB | | Aspartic acid, dipotassium salt | HMDB | | Aspartic acid, hydrobromide | HMDB | | Aspartic acid, monopotassium salt | HMDB | | Aspartic acid, monosodium salt | HMDB | | Aspartic acid, potassium salt | HMDB | | L Aspartate | HMDB | | MG5Longoral | HMDB | | Potassium aspartate | HMDB | | Polysuccinimide | HMDB | | Ammonium aspartate | HMDB | | Aspartate, ammonium | HMDB | | Aspartate, calcium | HMDB | | Aspartate, monosodium | HMDB | | Aspartic acid, calcium salt | HMDB | | Aspartic acid, disodium salt | HMDB | | Aspartic acid, magnesium (1:1) salt, hydrochloride, trihydrate | HMDB | | Dipotassium aspartate | HMDB | | Disodium aspartate | HMDB | | Hydrochloride, aspartate magnesium | HMDB | | Monopotassium aspartate | HMDB | | Sodium aspartate | HMDB | | Aspartate, dipotassium | HMDB | | Aspartic acid, magnesium (2:1) salt | HMDB | | Aspartic acid, sodium salt | HMDB | | Hydrobromide aspartic acid | HMDB | | Magnesium aspartate | HMDB | | MG 5 Longoral | HMDB | | Monosodium aspartate | HMDB | | Aspartate magnesium hydrochloride | HMDB | | Aspartate, potassium | HMDB | | Aspartate, sodium | HMDB | | Aspartic acid, ammonium salt | HMDB | | Aspartic acid, hydrochloride | HMDB | | Aspartic acid, magnesium-potassium (2:1:2) salt | HMDB | | Calcium aspartate | HMDB | | Hydrochloride aspartic acid | HMDB | | L Aspartic acid | HMDB | | Magnesiocard | HMDB | | MG-5-Longoral | HMDB | | Poly-DL-succinimide | HMDB |
|
|---|
| Chemical Formula | C4H7NO4 |
|---|
| Average Molecular Weight | 133.1 |
|---|
| Monoisotopic Molecular Weight | 133.0375 |
|---|
| IUPAC Name | (2S)-2-aminobutanedioic acid |
|---|
| Traditional Name | L-aspartic acid |
|---|
| CAS Registry Number | 56-84-8 |
|---|
| SMILES | N[C@@H](CC(O)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C4H7NO4/c5-2(4(8)9)1-3(6)7/h2H,1,5H2,(H,6,7)(H,8,9)/t2-/m0/s1 |
|---|
| InChI Key | CKLJMWTZIZZHCS-REOHCLBHSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as aspartic acid and derivatives. Aspartic acid and derivatives are compounds containing an aspartic acid or a derivative thereof resulting from reaction of aspartic acid at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Aspartic acid and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Aspartic acid or derivatives
- Alpha-amino acid
- L-alpha-amino acid
- Dicarboxylic acid or derivatives
- Fatty acid
- Amino acid
- Carboxylic acid
- Organic oxide
- Organopnictogen compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Organic oxygen compound
- Carbonyl group
- Amine
- Organic nitrogen compound
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 270 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 5.39 mg/mL | YALKOWSKY,SH & DANNENFELSER,RM (1992) | | logP | -3.89 | CHMELIK,J ET AL. (1991) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | Aspartic acid, 3 TMS, GC-MS Spectrum | splash10-0f89-0960000000-5a6a6cb21e9fc0875f84 | Spectrum | | GC-MS | Aspartic acid, 3 TMS, GC-MS Spectrum | splash10-0f89-0950000000-51399a8446c394459765 | Spectrum | | GC-MS | Aspartic acid, 3 TMS, GC-MS Spectrum | splash10-0f89-0950000000-f07e7f52b3c31fd119f5 | Spectrum | | GC-MS | Aspartic acid, 3 TMS, GC-MS Spectrum | splash10-0f89-0950000000-9e175dfa17a8b17a72d2 | Spectrum | | GC-MS | Aspartic acid, non-derivatized, GC-MS Spectrum | splash10-0f89-0950000000-f4475587d5a3b20bfc58 | Spectrum | | GC-MS | Aspartic acid, 3 TMS, GC-MS Spectrum | splash10-00di-9430000000-0cac9a97e698db1672b5 | Spectrum | | GC-MS | Aspartic acid, 2 TMS, GC-MS Spectrum | splash10-03yi-1900000000-ea61b8526ee02ae79b66 | Spectrum | | GC-MS | Aspartic acid, 3 TMS, GC-MS Spectrum | splash10-0f89-1890000000-7da576c8129142b71a1b | Spectrum | | GC-MS | Aspartic acid, 4 TMS, GC-MS Spectrum | splash10-0pb9-0981000000-ad23d55e348f55115f00 | Spectrum | | GC-MS | Aspartic acid, non-derivatized, GC-MS Spectrum | splash10-001i-0590000000-13cc5a7841854e97f68b | Spectrum | | GC-MS | Aspartic acid, non-derivatized, GC-MS Spectrum | splash10-0f89-0960000000-5a6a6cb21e9fc0875f84 | Spectrum | | GC-MS | Aspartic acid, non-derivatized, GC-MS Spectrum | splash10-0f89-0950000000-51399a8446c394459765 | Spectrum | | GC-MS | Aspartic acid, non-derivatized, GC-MS Spectrum | splash10-0f89-0950000000-f07e7f52b3c31fd119f5 | Spectrum | | GC-MS | Aspartic acid, non-derivatized, GC-MS Spectrum | splash10-0f89-0950000000-9e175dfa17a8b17a72d2 | Spectrum | | GC-MS | Aspartic acid, non-derivatized, GC-MS Spectrum | splash10-0f89-0950000000-f4475587d5a3b20bfc58 | Spectrum | | GC-MS | Aspartic acid, non-derivatized, GC-MS Spectrum | splash10-0g4i-6946100000-0ae49e3be0f2f1f92065 | Spectrum | | GC-MS | Aspartic acid, non-derivatized, GC-MS Spectrum | splash10-00di-9430000000-0cac9a97e698db1672b5 | Spectrum | | GC-MS | Aspartic acid, non-derivatized, GC-MS Spectrum | splash10-03yi-1900000000-ea61b8526ee02ae79b66 | Spectrum | | GC-MS | Aspartic acid, non-derivatized, GC-MS Spectrum | splash10-0f89-1890000000-7da576c8129142b71a1b | Spectrum | | GC-MS | Aspartic acid, non-derivatized, GC-MS Spectrum | splash10-0pb9-0981000000-ad23d55e348f55115f00 | Spectrum | | Predicted GC-MS | Aspartic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-000f-9100000000-b7db3377089f4c95001b | Spectrum | | Predicted GC-MS | Aspartic acid, 2 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-01w4-9610000000-0f1463c6395830708e12 | Spectrum | | Predicted GC-MS | Aspartic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Aspartic acid, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Aspartic acid, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0079-9300000000-92f9914d94078d96f5cf | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-00di-9000000000-c467d10f2b7e21ed1734 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00dl-9000000000-87f0c58226f5a2f3ac8a | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00di-0900000000-fde7ef1951fddff4b817 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0uk9-8900000000-2d7e5609618437e59272 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-4b43567f4a446aed0828 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-014i-4900000000-45382d9abd25be948e5b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-3f39a9b758e282358ac0 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-014i-3900000000-44d361ad09ff9a30dd14 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-18e1dba62e6b803e17b8 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0002-0930000000-d97f9518a2d516d830c5 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-02ai-0962100000-909bb894b2c1318afee4 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-9000000000-f2059dd438fcf62f62e1 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-03dr-7900000000-8b7b89ed34530e331024 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-014i-0190000000-58a78949f4b93de22aab | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-0921000000-214b6b969fdc4216ca71 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-03dr-6900000000-4918748927dd097d2879 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-0900000000-88dc2f1093f261e76201 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-0290000000-a8856f56645a2961baac | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-001i-0900000000-8cf9870557ca9adc3374 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-000i-9400000000-81b2804a712625a2d9c4 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-000i-9000000000-9bf77ba12ad952f84ac0 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-00di-9000000000-e08bfb96c5fd8c9b9db7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0006-9000000000-876e30f6c9ed061091fe | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-00dl-9000000000-6c0d44d4e3853e5701a3 | 2012-08-31 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 400 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Transcription/Translation | Not Available | Not Available | | Arginine and Proline Metabolism | 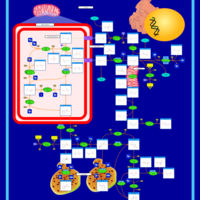 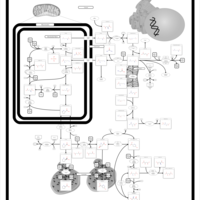 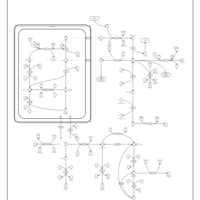 |  | | Aspartate Metabolism | 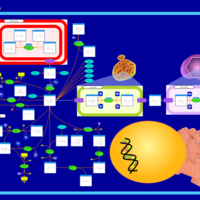   |  | | Urea Cycle |   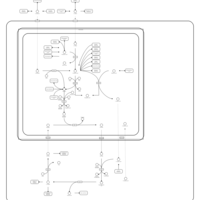 |  | | Beta-Alanine Metabolism | 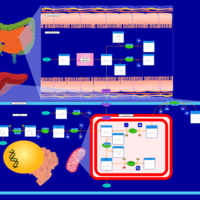 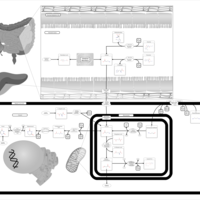 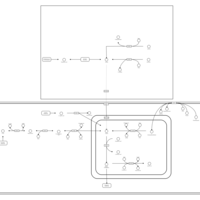 |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
| Calcium-binding mitochondrial carrier protein Aralar2 | SLC25A13 | 7q21.3 | Q9UJS0 | details | | Excitatory amino acid transporter 3 | SLC1A1 | 9p24 | P43005 | details | | Calcium-binding mitochondrial carrier protein Aralar1 | SLC25A12 | 2q24 | O75746 | details | | Monocarboxylate transporter 10 | SLC16A10 | 6q21-q22 | Q8TF71 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 1.446 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 1.278 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 0.827 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.391 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 1.609 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 0.675 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000191 |
|---|
| DrugBank ID | DB00128 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB012567 |
|---|
| KNApSAcK ID | C00001342 |
|---|
| Chemspider ID | 5745 |
|---|
| KEGG Compound ID | C00049 |
|---|
| BioCyc ID | L-ASPARTATE |
|---|
| BiGG ID | 33663 |
|---|
| Wikipedia Link | Aspartic acid |
|---|
| METLIN ID | 5206 |
|---|
| PubChem Compound | 5960 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17053 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
|
|---|


