| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:26:25 UTC |
|---|
| Updated at | 2020-12-07 19:07:10 UTC |
|---|
| CannabisDB ID | CDB000144 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Myoinositol |
|---|
| Description | Myo-Inositol is an inositol isoform. Inositol is a derivative of cyclohexane with six hydroxyl groups, making it a polyol. It also is known as a sugar alcohol, having the same molecular formula as glucose or other hexoses. Inositol exists in nine possible stereoisomers, including scyllo-inositol, myo-inositol (the most abundant), muco-inositol, D-chiro-inositol, L-chiro-inositol, neo-inositol, allo-inositol, epi-inositol, and cis-inositol. In humans, most inositol is synthesized in the kidneys, typically in amounts of a few grams per day. It is found in many foods, particularly in cereals with high bran content. Inositol is one of the sugar alcohols and cyclitols identified in Cannabis sativa plants (PMID: 6991645 ). It is an isomer of glucose that has traditionally been a B vitamin although it has an uncertain status as a vitamin and a deficiency syndrome has not been identified in humans. Inositol is a cyclic polyalcohol that plays an important role as a second messenger in a cell, in the form of inositol phosphates. Inositol phospholipids are important in signal transduction. A possible health effect of inositols, including myo-inositol, was recently reviewed. The review concluded that myo-inositol, together with chiro-inositol, at a 40:1 ratio, were an important therapeutic strategy for the improvement of metabolic, hormonal, and reproductive aspects of polycystic ovarian syndrome (PMID: 29309199 ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (1R,2R,3S,4S,5R,6S)-Cyclohexane-1,2,3,4,5,6-hexol | ChEBI | | 1,2,3,4,5,6-HEXAHYDROXY-cyclohexane | ChEBI | | 1,2,3,5/4,6-cyclohexanehexol | ChEBI | | 1D-Myo-inositol | ChEBI | | 1l-Myo-inositol | ChEBI | | Bios I | ChEBI | | cis-1,2,3,5-trans-4,6-Cyclohexanehexol | ChEBI | | Cyclohexitol | ChEBI | | D-Myo-inositol | ChEBI | | Dambose | ChEBI | | I-inositol | ChEBI | | Inosite | ChEBI | | Inositol | ChEBI | | Ins | ChEBI | | L-Myo-inositol | ChEBI | | Meat sugar | ChEBI | | Meso-inositol | ChEBI | | Myoinositol | ChEBI | | Inosital | Kegg | | Iso-inositol | HMDB | | MI | HMDB | | Myoinosite | HMDB | | Phaseomannite | HMDB | | Rat antispectacled eye factor | HMDB | | Myo inositol | HMDB | | Vitamin b8 | HMDB | | Myo-inositol | HMDB |
|
|---|
| Chemical Formula | C6H12O6 |
|---|
| Average Molecular Weight | 180.16 |
|---|
| Monoisotopic Molecular Weight | 180.0634 |
|---|
| IUPAC Name | (1R,2R,3r,4S,5S,6s)-cyclohexane-1,2,3,4,5,6-hexol |
|---|
| Traditional Name | L-inositol |
|---|
| CAS Registry Number | 87-89-8 |
|---|
| SMILES | O[C@H]1[C@H](O)[C@@H](O)[C@H](O)[C@H](O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C6H12O6/c7-1-2(8)4(10)6(12)5(11)3(1)9/h1-12H/t1-,2-,3-,4+,5-,6- |
|---|
| InChI Key | CDAISMWEOUEBRE-GPIVLXJGSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as cyclohexanols. Cyclohexanols are compounds containing an alcohol group attached to a cyclohexane ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Alcohols and polyols |
|---|
| Direct Parent | Cyclohexanols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Cyclohexanol
- Sugar alcohol
- Cyclitol or derivatives
- Cyclic alcohol
- Polyol
- Hydrocarbon derivative
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 225 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 143 mg/mL at 19 °C | Human Metabolome Project | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | Myoinositol, 6 TMS, GC-MS Spectrum | splash10-014j-0953000000-1571a0577293e96091b1 | Spectrum | | GC-MS | Myoinositol, 6 TMS, GC-MS Spectrum | splash10-00kb-0932000000-96ddc21293431d746ad2 | Spectrum | | GC-MS | Myoinositol, non-derivatized, GC-MS Spectrum | splash10-00kb-0942000000-b78076224adf13bade25 | Spectrum | | GC-MS | Myoinositol, non-derivatized, GC-MS Spectrum | splash10-00kb-0942000000-42c4e40d04b4f306535d | Spectrum | | GC-MS | Myoinositol, 6 TMS, GC-MS Spectrum | splash10-00di-8942000000-f5d7abd38d6ff547a0a9 | Spectrum | | GC-MS | Myoinositol, 6 TMS, GC-MS Spectrum | splash10-066r-0975000000-df7304c12b6c9bdb0f76 | Spectrum | | GC-MS | Myoinositol, non-derivatized, GC-MS Spectrum | splash10-014j-0953000000-1571a0577293e96091b1 | Spectrum | | GC-MS | Myoinositol, non-derivatized, GC-MS Spectrum | splash10-00kb-0932000000-96ddc21293431d746ad2 | Spectrum | | GC-MS | Myoinositol, non-derivatized, GC-MS Spectrum | splash10-00kb-0942000000-b78076224adf13bade25 | Spectrum | | GC-MS | Myoinositol, non-derivatized, GC-MS Spectrum | splash10-00di-8942000000-f5d7abd38d6ff547a0a9 | Spectrum | | GC-MS | Myoinositol, non-derivatized, GC-MS Spectrum | splash10-066r-0975000000-df7304c12b6c9bdb0f76 | Spectrum | | Predicted GC-MS | Myoinositol, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-03dr-9700000000-18a13ab4150e6ebb20cd | Spectrum | | Predicted GC-MS | Myoinositol, 6 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00fr-9001040000-00e67b437c4329aee95d | Spectrum | | Predicted GC-MS | Myoinositol, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Myoinositol, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Myoinositol, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Myoinositol, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Myoinositol, TMS_1_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Myoinositol, TMS_1_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Myoinositol, TMS_1_6, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Myoinositol, TMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Myoinositol, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Myoinositol, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Myoinositol, TMS_2_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Myoinositol, TMS_2_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, N/A (Annotated) | splash10-0a59-3900000000-96084971c3e13a1e7451 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, N/A (Annotated) | splash10-001i-9000000000-1de17d7f53ca727e6d72 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, N/A (Annotated) | splash10-0zh9-9000000000-bef910214341056a78aa | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 2V, negative | splash10-004i-0900000000-41a49af846b1636763b5 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 2V, negative | splash10-004i-0900000000-06f9ecfccf30d40ef204 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 3V, negative | splash10-004i-0900000000-bca88f6782a8ed386223 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 4V, negative | splash10-004i-1900000000-500ab195d69568b8678b | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 5V, negative | splash10-01ti-5900000000-a1657b54c985e2be1d19 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 6V, negative | splash10-000i-9600000000-325cbb8dbfdeafb2b31f | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 8V, negative | splash10-000i-9200000000-9145a4766351204e723a | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 9V, negative | splash10-000i-9100000000-2bbbcfa4733b749ec768 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 10V, negative | splash10-000i-9000000000-de0ec8731a152b612bb9 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 12V, negative | splash10-0079-9000000000-41d049d1feef94e190d4 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 14V, negative | splash10-059i-9000000000-796e728daf28d5122dee | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 15V, negative | splash10-059i-9000000000-8643e206319f3008a8d4 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 18V, negative | splash10-0abi-9000000000-98e61f19bc2842f2e97b | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 20V, negative | splash10-0ab9-9000000000-29aa062fc4d5341167b6 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 12V, negative | splash10-03di-1900000000-5a2897fd032a959918b3 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 12V, negative | splash10-000i-9000000000-71c4786b7034d5a0a0a9 | 2020-07-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0900000000-5cb39f92646251665ebf | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0900000000-2aeb7708f31c772e2d95 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08gi-8900000000-74f89711a1c6891808a4 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0900000000-8b68ff47f846a8ca1ddd | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-1900000000-03ef8572289795275d9c | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0570-9300000000-950a37bd331028801e9f | 2016-09-12 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Inositol Metabolism | 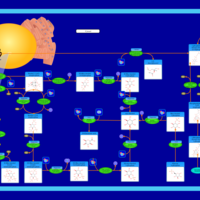 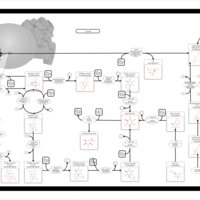 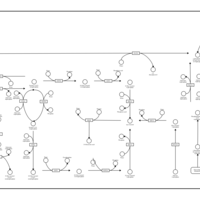 |  | | Galactose Metabolism |  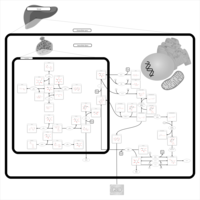 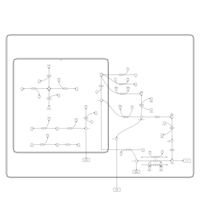 |  | | Phosphatidylinositol Phosphate Metabolism | 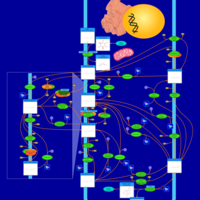 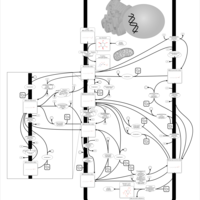 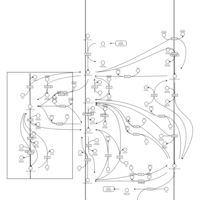 |  | | Inositol Phosphate Metabolism | 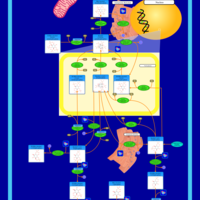 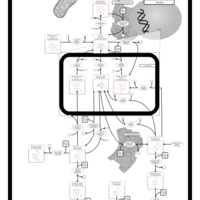 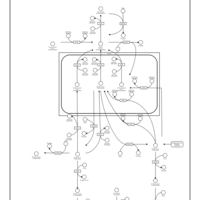 |  | | Joubert syndrome | 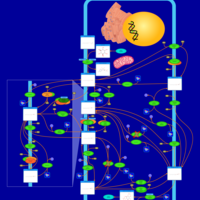 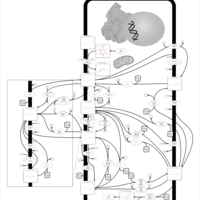 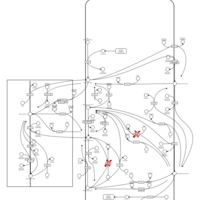 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000211 |
|---|
| DrugBank ID | DB13178 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB010547 |
|---|
| KNApSAcK ID | C00001164 |
|---|
| Chemspider ID | 10239179 |
|---|
| KEGG Compound ID | C00137 |
|---|
| BioCyc ID | MYO-INOSITOL |
|---|
| BiGG ID | 33990 |
|---|
| Wikipedia Link | Inositol |
|---|
| METLIN ID | 5221 |
|---|
| PubChem Compound | Not Available |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17268 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
- Gateva A, Unfer V, Kamenov Z: The use of inositol(s) isomers in the management of polycystic ovary syndrome: a comprehensive review. Gynecol Endocrinol. 2018 Jul;34(7):545-550. doi: 10.1080/09513590.2017.1421632. Epub 2018 Jan 8. [PubMed:29309199 ]
|
|---|

