| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:26:03 UTC |
|---|
| Updated at | 2022-12-13 23:36:24 UTC |
|---|
| CannabisDB ID | CDB000134 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | L-Phenylalanine |
|---|
| Description | Phenylalanine, abbreviated Phe or F, is a neutral, nonpolar, hydrophobic, essential amino acid. It is the precursor of the amino acid tyrosine, a precursor of the skin pigment melanin and a precursor of catecholamines that include tyramine, dopamine, epinephrine, and norepinephrine. Catecholamines are neurotransmitters that act as adrenalin-like substances. Interestingly, several psychotropic drugs (mescaline, morphine, codeine, and papaverine) also have phenylalanine as a constituent. Phenylalanine is highly concentrated in the human brain and plasma and is found naturally in mammalian breast milk. Normal metabolism of phenylalanine requires biopterin, iron, niacin, vitamin B6, copper, and vitamin C. An average adult ingests 5 g of phenylalanine per day and may optimally need up to 8 g daily. Phenylalanine is highly concentrated in several high protein foods, such as meat, cottage cheese, and wheat germ. Phenylalanine is also found in Cannabis plants (PMID: 6991645 ). An additional dietary source of phenylalanine is artificial sweeteners containing aspartame. Generally, aspartame should be avoided by phenylketonurics and pregnant women. When present in sufficiently high levels, phenylalanine can act as a neurotoxin and a metabotoxin. A neurotoxin is a compound that disrupts or attacks neural cells and neural tissue. A metabotoxin is an endogenously produced metabolite that causes adverse health effects at chronically high levels. Chronically high levels of phenylalanine are associated with at least five inborn errors of metabolism, including Hartnup disorder, hyperphenylalaninemia due to guanosine triphosphate cyclohydrolase deficiency, phenylketonuria (PKU), tyrosinemia type 2 (or Richner-Hanhart syndrome), and tyrosinemia type III (TYRO3). Untreated PKU can lead to intellectual disability, seizures, behavioural problems, and mental disorders. Classic PKU dramatically affects myelination and white matter tracts in untreated infants; this may be one major cause of neurological disorders associated with PKU. PKU patients have elevated serum plasma levels of phenylalanine, up to 400 times normal. High plasma concentrations of phenylalanine influence the blood-brain barrier transport of large neutral amino acids. The high plasma phenylalanine concentrations increase phenylalanine entry into the brain and restrict the entry of other large neutral amino acids (PMID: 19191004 ). Phenylalanine has been found to interfere with different cerebral enzyme systems. PKU individuals may have a musty smell and lighter skin, likely related to altered conversion of phenylalanine to the skin pigment melanin. Mild phenylketonuria may be an unsuspected cause of hyperactivity, learning problems, and other developmental problems in children. It has been recently suggested that PKU may resemble amyloid diseases, such as Alzheimer's disease and Parkinson's disease, due to the formation of toxic amyloid-like assemblies of phenylalanine (PMID: 22706200 ). Phenylalanine also has some potential benefits. Phenylalanine can act as an analgesic, providing relief for premenstrual syndrome and may enhance the effects of acupuncture and electric transcutaneous nerve stimulation (TENS) for Parkinson's patients. Phenylalanine and tyrosine, like L-DOPA, produce a catecholamine-like effect. Phenylalanine is better absorbed than tyrosine and may cause fewer headaches. Low phenylalanine diets have been prescribed for certain cancers with mixed results. For some tumours that use more phenylalanine than others (such as melatonin-producing tumours called melanomas) may benefit from low phenylalanine diets. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (S)-2-Amino-3-phenylpropionic acid | ChEBI | | (S)-alpha-Amino-beta-phenylpropionic acid | ChEBI | | 3-Phenyl-L-alanine | ChEBI | | beta-Phenyl-L-alanine | ChEBI | | F | ChEBI | | Phe | ChEBI | | PHENYLALANINE | ChEBI | | (S)-2-Amino-3-phenylpropionate | Generator | | (S)-a-Amino-b-phenylpropionate | Generator | | (S)-a-Amino-b-phenylpropionic acid | Generator | | (S)-alpha-Amino-beta-phenylpropionate | Generator | | (S)-Α-amino-β-phenylpropionate | Generator | | (S)-Α-amino-β-phenylpropionic acid | Generator | | b-Phenyl-L-alanine | Generator | | Β-phenyl-L-alanine | Generator | | (-)-beta-Phenylalanine | HMDB | | (L)-Phenylalanine | HMDB | | (S)-(-)-Phenylalanine | HMDB | | (S)-2-Amino-3-phenylpropanoate | HMDB | | (S)-2-Amino-3-phenylpropanoic acid | HMDB | | (S)-alpha-Amino-benzenepropanoate | HMDB | | (S)-alpha-Amino-benzenepropanoic acid | HMDB | | (S)-alpha-Aminobenzenepropanoate | HMDB | | (S)-alpha-Aminobenzenepropanoic acid | HMDB | | (S)-alpha-Aminohydrocinnamate | HMDB | | (S)-alpha-Aminohydrocinnamic acid | HMDB | | (S)-Phenylalanine | HMDB | | alpha-Aminohydrocinnamate | HMDB | | alpha-Aminohydrocinnamic acid | HMDB | | beta-Phenyl-alpha-alanine | HMDB | | beta-Phenylalanine | HMDB | | L-2-Amino-3-phenylpropionate | HMDB | | L-2-Amino-3-phenylpropionic acid | HMDB | | Phenyl-alanine | HMDB | | Phenylalamine | HMDB | | L-Isomer phenylalanine | HMDB | | Phenylalanine, L isomer | HMDB | | Phenylalanine, L-isomer | HMDB | | Endorphenyl | HMDB | | (6S)-Tetrahydrofolate | HMDB | | (6S)-Tetrahydrofolic acid | HMDB | | (6S)-THFA | HMDB | | 5,6,7,8-Tetrahydrofolate | HMDB | | Tetrahydrofolate | HMDB | | THF | HMDB | | 5,6,7,8-Tetrahydrofolic acid | HMDB |
|
|---|
| Chemical Formula | C9H11NO2 |
|---|
| Average Molecular Weight | 165.19 |
|---|
| Monoisotopic Molecular Weight | 165.079 |
|---|
| IUPAC Name | (2S)-2-amino-3-phenylpropanoic acid |
|---|
| Traditional Name | L-phenylalanine |
|---|
| CAS Registry Number | 5297-02-9 |
|---|
| SMILES | N[C@@H](CC1=CC=CC=C1)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C9H11NO2/c10-8(9(11)12)6-7-4-2-1-3-5-7/h1-5,8H,6,10H2,(H,11,12)/t8-/m0/s1 |
|---|
| InChI Key | COLNVLDHVKWLRT-QMMMGPOBSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as phenylalanine and derivatives. Phenylalanine and derivatives are compounds containing phenylalanine or a derivative thereof resulting from reaction of phenylalanine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Phenylalanine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Phenylalanine or derivatives
- 3-phenylpropanoic-acid
- Alpha-amino acid
- Amphetamine or derivatives
- L-alpha-amino acid
- Aralkylamine
- Monocyclic benzene moiety
- Benzenoid
- Amino acid
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic nitrogen compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Hydrocarbon derivative
- Primary aliphatic amine
- Organic oxide
- Carbonyl group
- Organopnictogen compound
- Organic oxygen compound
- Amine
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Indirect biological role: Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 283 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 26.9 mg/mL | Not Available | | logP | -1.38 | AVDEEF,A (1997) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-00dl-9300000000-4782928378caea601f9b | 2018-05-25 | View Spectrum | | GC-MS | L-Phenylalanine, 2 TMS, GC-MS Spectrum | splash10-0fr6-1930000000-a37fbccaf826443ef70c | Spectrum | | GC-MS | L-Phenylalanine, non-derivatized, GC-MS Spectrum | splash10-014l-1940000000-537e2725d621246630c1 | Spectrum | | GC-MS | L-Phenylalanine, 2 TMS, GC-MS Spectrum | splash10-00di-9630000000-ead0919f9a19d2352d80 | Spectrum | | GC-MS | L-Phenylalanine, 1 TMS, GC-MS Spectrum | splash10-00di-2900000000-cb2d6dc4bf9515150328 | Spectrum | | GC-MS | L-Phenylalanine, 2 TMS, GC-MS Spectrum | splash10-014l-2960000000-f77489792f0652dd5613 | Spectrum | | GC-MS | L-Phenylalanine, non-derivatized, GC-MS Spectrum | splash10-014l-0970000000-792b341dd28b9e30bac2 | Spectrum | | GC-MS | L-Phenylalanine, non-derivatized, GC-MS Spectrum | splash10-0fr6-1930000000-a37fbccaf826443ef70c | Spectrum | | GC-MS | L-Phenylalanine, non-derivatized, GC-MS Spectrum | splash10-014l-1940000000-537e2725d621246630c1 | Spectrum | | GC-MS | L-Phenylalanine, non-derivatized, GC-MS Spectrum | splash10-0ul9-3659000000-b85be4e71798e2fdc47b | Spectrum | | GC-MS | L-Phenylalanine, non-derivatized, GC-MS Spectrum | splash10-00di-9630000000-ead0919f9a19d2352d80 | Spectrum | | GC-MS | L-Phenylalanine, non-derivatized, GC-MS Spectrum | splash10-00di-2900000000-cb2d6dc4bf9515150328 | Spectrum | | GC-MS | L-Phenylalanine, non-derivatized, GC-MS Spectrum | splash10-014l-2960000000-f77489792f0652dd5613 | Spectrum | | Predicted GC-MS | L-Phenylalanine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-006x-9600000000-df38fcb743d8f44fb876 | Spectrum | | Predicted GC-MS | L-Phenylalanine, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00di-7900000000-f21569d2ec75b88e1bda | Spectrum | | Predicted GC-MS | L-Phenylalanine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Phenylalanine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Phenylalanine, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Phenylalanine, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Phenylalanine, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-014i-0900000000-0f3b994108b8a9fd2a56 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0gb9-2900000000-c14d44c8a67621757f3d | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-004i-9300000000-08c642dab7f49c00da43 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-014i-0900000000-7dce1e473976f7d2143e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0f6x-9600000000-711557391093b0d8500a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00di-0900000000-0c25a5c116eac7bb059b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00di-0900000000-2804f79084ac4e67e155 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-014i-0900000000-8a71bb1f8424064d7caf | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0f6x-9800000000-e027ff6bb67ce55e80a5 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-014i-0900000000-9f6185e9c7d54189f369 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00di-0900000000-df5f72fe2bba91742427 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0ir3-0988735721-bac229222fe7b52812a8 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0002-0900000000-453477dec847a3672ffe | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0a4i-0010963000-079a66bf710f6778bceb | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-0000009000-62dbe98de4ecde484fb3 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-03di-0900000000-80558c17dc1845663c85 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0002-3900000000-e1ee31d41e48824e84b7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0f6x-9500000000-cc11290a37615f24e16e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0006-9000000000-1a6020bce0e1a9a14832 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0006-9000000000-38c044a112152626962e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-014i-0900000000-9b908abfcb63153d60b3 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-00di-1900000000-3ba5964e151bb1d56188 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-00di-2900000000-20a7d24da0281f5b3b78 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-0udi-5900000000-1800642a835b49f3398a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-0fb9-9300000000-f262384c85fb843f8a11 | 2012-08-31 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Transcription/Translation | Not Available | Not Available | | Phenylalanine and Tyrosine Metabolism | 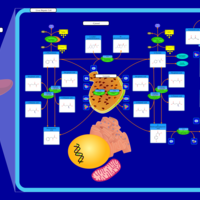 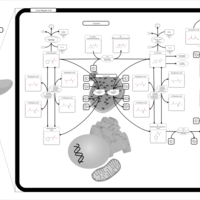 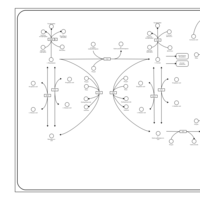 |  | | Phenylketonuria | 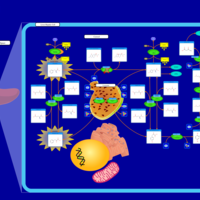 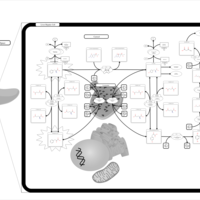 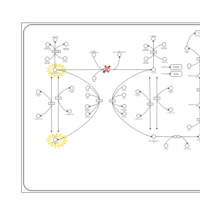 | Not Available | | Tyrosinemia Type 2 (or Richner-Hanhart syndrome) | 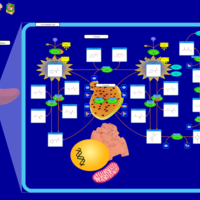 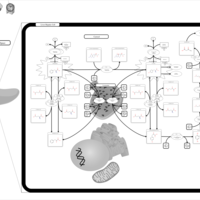 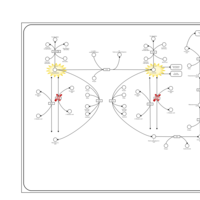 | Not Available | | Tyrosinemia Type 3 (TYRO3) | 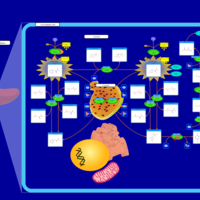 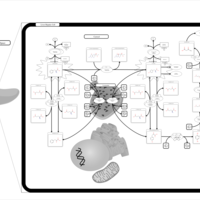 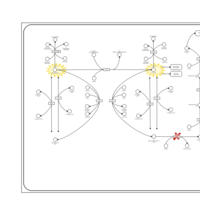 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
| Large neutral amino acids transporter small subunit 2 | SLC7A8 | 14q11.2 | Q9UHI5 | details | | Monocarboxylate transporter 10 | SLC16A10 | 6q21-q22 | Q8TF71 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 0.307 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 0.335 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 0.150 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.208 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 0.143 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 0.0507 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000159 |
|---|
| DrugBank ID | DB00120 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB004940 |
|---|
| KNApSAcK ID | C00001386 |
|---|
| Chemspider ID | 5910 |
|---|
| KEGG Compound ID | C00079 |
|---|
| BioCyc ID | PHE |
|---|
| BiGG ID | 33775 |
|---|
| Wikipedia Link | Phenylalanine |
|---|
| METLIN ID | 28 |
|---|
| PubChem Compound | 6140 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17295 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
- van Spronsen FJ, Hoeksma M, Reijngoud DJ: Brain dysfunction in phenylketonuria: is phenylalanine toxicity the only possible cause? J Inherit Metab Dis. 2009 Feb;32(1):46-51. doi: 10.1007/s10545-008-0946-2. Epub 2009 Jan 13. [PubMed:19191004 ]
- Adler-Abramovich L, Vaks L, Carny O, Trudler D, Magno A, Caflisch A, Frenkel D, Gazit E: Phenylalanine assembly into toxic fibrils suggests amyloid etiology in phenylketonuria. Nat Chem Biol. 2012 Aug;8(8):701-6. doi: 10.1038/nchembio.1002. Epub 2012 Jun 17. [PubMed:22706200 ]
|
|---|
