| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:24:57 UTC |
|---|
| Updated at | 2022-12-13 23:36:22 UTC |
|---|
| CannabisDB ID | CDB000105 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Proline |
|---|
| Description | Proline or L-proline abbreviated Pro or is one of the twenty amino acids used in living organisms as the building blocks of proteins. Proline is sometimes called an imino acid, although the IUPAC definition of an imine requires a carbon-nitrogen double bond. Proline is a non-essential amino acid that is largely synthesized from glutamic acid. While proline biosynthetic pathways exist from eubacteria to eukaryotes, in the most prevalent pathway, proline is cyclized from glutamate. First glutamate is phosphorylated to gamma-glutamyl phosphate by gamma-glutamyl kinase, then second, reduced to gamma-glutamyl semialdehyde by gamma-glutamyl phosphate reductase, then third, cyclized spontaneously to delta(1)-pyrroline-5-carboxylate and fourth, reduced to proline by delta(1)-pyrroline-5-carboxylate reductase. In higher plants and animals, the first two steps are catalysed by a bi-functional delta(1) -pyrroline-5-carboxylate synthase. Alternative pathways of proline formation use the initial steps of the arginine biosynthetic pathway to ornithine, which can be converted to delta(1)-pyrroline-5-carboxylate by ornithine aminotransferase and then reduced to proline or converted directly to proline by ornithine cyclodeaminase (PMID: 25367752 ). Proline is an essential component of collagen and is important for proper functioning of joints and tendons. L-proline has acted as a weak agonist of the glycine receptor and of both NMDA and non-NMDA ionotropic glutamate receptors (PMID:1349155 ). It is a potential endogenous excitotoxin/neurotoxin, which causes damage to nerve cells and nerve tissues. Proline, when injected into the brains of rats, non-selectively destroyed pyramidal and granule cells (PMID: 3409032 ) suggesting that it can act as a neurotoxin. Proline can also be a metabotoxin which is an endogenously produced metabolite that causes adverse health effects at chronically high levels. At least five inborn errors of metabolism, including hyperprolinemia type I, hyperprolinemia type II, iminoglycinuria, prolinemia type II, and pyruvate carboxylase deficiency are associated with chronically high levels of proline (PMID: 18806117 ). While most people with hyperprolinemia type I often do not show any symptoms despite proline blood levels being between 3 and 10 times the normal level, some individuals have seizures, intellectual disability, or other neurological or psychiatric problems. Hyperprolinemia type II patients have proline blood levels between 10 and 15 times higher than normal, and high levels of a related compound called pyrroline-5-carboxylate. While Hyperprolinemia type II patients have signs and symptoms that vary in severity, they are more likely than type I patients to have seizures or intellectual disability. Hyperprolinemia I and II are caused by deficiencies in the enzymatic activities of proline dehydrogenase and Delta-1-pyrroline-5-carboxylate reductase, respectively. Proline is also found in Cannabis plants (PMID: 6991645 ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (-)-(S)-Proline | ChEBI | | (-)-2-Pyrrolidinecarboxylic acid | ChEBI | | (-)-Proline | ChEBI | | (2S)-Pyrrolidine-2-carboxylic acid | ChEBI | | (S)-2-Carboxypyrrolidine | ChEBI | | (S)-2-Pyrrolidinecarboxylic acid | ChEBI | | (S)-Pyrrolidine-2-carboxylic acid | ChEBI | | 2-Pyrrolidinecarboxylic acid | ChEBI | | L-(-)-Proline | ChEBI | | L-alpha-Pyrrolidinecarboxylic acid | ChEBI | | L-Prolin | ChEBI | | L-Pyrrolidine-2-carboxylic acid | ChEBI | | P | ChEBI | | Prolina | ChEBI | | PROLINE | ChEBI | | Prolinum | ChEBI | | (-)-2-Pyrrolidinecarboxylate | Generator | | (2S)-Pyrrolidine-2-carboxylate | Generator | | (S)-2-Pyrrolidinecarboxylate | Generator | | (S)-Pyrrolidine-2-carboxylate | Generator | | 2-Pyrrolidinecarboxylate | Generator | | L-a-Pyrrolidinecarboxylate | Generator | | L-a-Pyrrolidinecarboxylic acid | Generator | | L-alpha-Pyrrolidinecarboxylate | Generator | | L-Α-pyrrolidinecarboxylate | Generator | | L-Α-pyrrolidinecarboxylic acid | Generator | | L-Pyrrolidine-2-carboxylate | Generator | | (S)-(-)-Proline | HMDB | | (S)-(-)-Pyrrolidine-2-carboxylate | HMDB | | (S)-(-)-Pyrrolidine-2-carboxylic acid | HMDB | | (S)-2-Pyrralidinecarboxylate | HMDB | | (S)-2-Pyrralidinecarboxylic acid | HMDB | | (S)-Proline | HMDB | | L Proline | HMDB |
|
|---|
| Chemical Formula | C5H9NO2 |
|---|
| Average Molecular Weight | 115.13 |
|---|
| Monoisotopic Molecular Weight | 115.0633 |
|---|
| IUPAC Name | (2S)-pyrrolidine-2-carboxylic acid |
|---|
| Traditional Name | L-proline |
|---|
| CAS Registry Number | 147-85-3 |
|---|
| SMILES | OC(=O)[C@@H]1CCCN1 |
|---|
| InChI Identifier | InChI=1S/C5H9NO2/c7-5(8)4-2-1-3-6-4/h4,6H,1-3H2,(H,7,8)/t4-/m0/s1 |
|---|
| InChI Key | ONIBWKKTOPOVIA-BYPYZUCNSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as proline and derivatives. Proline and derivatives are compounds containing proline or a derivative thereof resulting from reaction of proline at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Proline and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Proline or derivatives
- Alpha-amino acid
- L-alpha-amino acid
- Pyrrolidine carboxylic acid
- Pyrrolidine carboxylic acid or derivatives
- Pyrrolidine
- Amino acid
- Carboxylic acid
- Secondary aliphatic amine
- Monocarboxylic acid or derivatives
- Secondary amine
- Organoheterocyclic compound
- Azacycle
- Organic oxygen compound
- Organooxygen compound
- Organonitrogen compound
- Amine
- Organopnictogen compound
- Organic oxide
- Carbonyl group
- Organic nitrogen compound
- Hydrocarbon derivative
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Indirect biological role: Environmental role: Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 221 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 162 mg/mL | Not Available | | logP | -2.54 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | Proline, 2 TMS, GC-MS Spectrum | splash10-0006-0910000000-a9127c0e370afb80259b | Spectrum | | GC-MS | Proline, 2 TMS, GC-MS Spectrum | splash10-000f-0910000000-3195f21e625520d51316 | Spectrum | | GC-MS | Proline, non-derivatized, GC-MS Spectrum | splash10-0006-0900000000-119af746c347969e8ae7 | Spectrum | | GC-MS | Proline, 2 TMS, GC-MS Spectrum | splash10-006x-8900000000-b31beabb1ed6235e7145 | Spectrum | | GC-MS | Proline, 1 TMS, GC-MS Spectrum | splash10-00di-9100000000-c1c67bc521741ee891cb | Spectrum | | GC-MS | Proline, 2 TMS, GC-MS Spectrum | splash10-0006-1900000000-84e6fb318323fd0261b1 | Spectrum | | GC-MS | Proline, 1 TMS, GC-MS Spectrum | splash10-00xs-8910000000-74b0c34ca5fd7c85f1d9 | Spectrum | | GC-MS | Proline, non-derivatized, GC-MS Spectrum | splash10-00fu-9000000000-716c3d01b301162863b6 | Spectrum | | GC-MS | Proline, non-derivatized, GC-MS Spectrum | splash10-0006-0900000000-0d7354201ed866c87b13 | Spectrum | | GC-MS | Proline, non-derivatized, GC-MS Spectrum | splash10-0006-0910000000-a9127c0e370afb80259b | Spectrum | | GC-MS | Proline, non-derivatized, GC-MS Spectrum | splash10-000f-0910000000-3195f21e625520d51316 | Spectrum | | GC-MS | Proline, non-derivatized, GC-MS Spectrum | splash10-0006-0900000000-119af746c347969e8ae7 | Spectrum | | GC-MS | Proline, non-derivatized, GC-MS Spectrum | splash10-001i-1920000000-09006ee4e7a137d8a45a | Spectrum | | GC-MS | Proline, non-derivatized, GC-MS Spectrum | splash10-006x-8900000000-b31beabb1ed6235e7145 | Spectrum | | GC-MS | Proline, non-derivatized, GC-MS Spectrum | splash10-0006-1900000000-84e6fb318323fd0261b1 | Spectrum | | GC-MS | Proline, non-derivatized, GC-MS Spectrum | splash10-00xs-8910000000-74b0c34ca5fd7c85f1d9 | Spectrum | | GC-MS | Proline, non-derivatized, GC-MS Spectrum | splash10-00di-9100000000-c1c67bc521741ee891cb | Spectrum | | Predicted GC-MS | Proline, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00fu-9000000000-d203e3dfb0701403a75b | Spectrum | | Predicted GC-MS | Proline, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00di-9000000000-56471980a375d26046ca | Spectrum | | Predicted GC-MS | Proline, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Proline, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Proline, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Proline, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-00xr-9600000000-ba51a901e786b1a4663e | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-00di-9000000000-f72e2f3812225708fc56 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00di-9000000000-59887da6580f63bcc1c5 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI M-80) , Positive | splash10-00fu-9000000000-c69dcaf7cb2d1e9d285e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-014i-0930000000-984f9a23ebfe28f95017 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-014i-9000000000-f75575cac28a54aa923d | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00di-9000000000-221cfb8c82f3df76eb51 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-014i-0900000000-e5741fd9870218d57caf | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-014i-0900000000-6bc9b4e775be37fa18d9 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-014i-9000000000-a8eb419b26655a6a1dab | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00di-9000000000-873ee03b61d1ffcae70f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-014i-0900000000-61d583960651669fb9e6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-03di-0900000000-2c23774af5f6a0c217d5 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-03di-0900000000-d89c1e626ef24a79c89f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-03di-1900000000-fc27094f2eb75e65b438 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-014i-1900000000-3221c23504b8847abf8d | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-00di-9100000000-8169367f5127c57fae12 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-00di-9000000000-f704eb8402fcacfccd18 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-00di-9000000000-ccd74eea36efab2cbab8 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-00di-9000000000-9f214692cd39a6581813 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - CE-ESI-TOF (CE-system connected to 6210 Time-of-Flight MS, Agilent) , Positive | splash10-014i-0900000000-ef038c77922e4c89eeb0 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-00di-9400000000-4073ea1df57a60b36d69 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) 30V, Positive | splash10-00di-9000000000-495fdbd2f565bd794632 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-03di-0900000000-e1d0a4b20419dc7bae92 | 2012-08-31 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-6900000000-eba8ceb2a71d2726acd0 | 2016-09-12 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Transcription/Translation | Not Available | Not Available | | Arginine and Proline Metabolism | 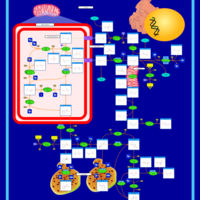 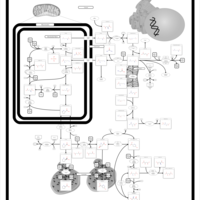 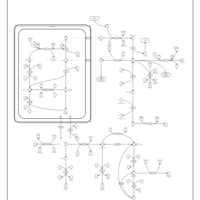 |  | | Prolidase Deficiency (PD) | 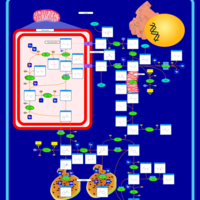 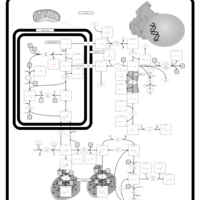 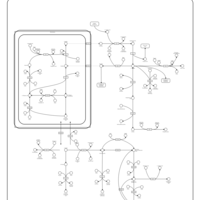 | Not Available | | Arginine: Glycine Amidinotransferase Deficiency (AGAT Deficiency) | 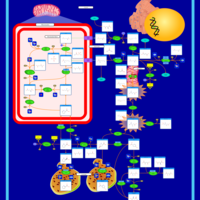 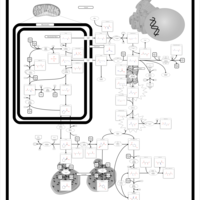  | Not Available | | Hyperprolinemia Type II |    | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
| Glutamate [NMDA] receptor subunit zeta-1 | GRIN1 | 9q34.3 | Q05586 | details | | Glutamate [NMDA] receptor subunit 3B | GRIN3B | 19p13.3 | O60391 | details | | Glutamate [NMDA] receptor subunit epsilon-1 | GRIN2A | 16p13.2 | Q12879 | details | | Glutamate [NMDA] receptor subunit epsilon-3 | GRIN2C | 17q25 | Q14957 | details | | Glutamate [NMDA] receptor subunit epsilon-2 | GRIN2B | 12p12 | Q13224 | details | | Glutamate [NMDA] receptor subunit epsilon-4 | GRIN2D | 19q13.1-qter | O15399 | details | | Sodium-dependent proline transporter | SLC6A7 | 5q32 | Q99884 | details | | Sodium- and chloride-dependent neutral and basic amino acid transporter B(0+) | SLC6A14 | | Q9UN76 | details | | Monocarboxylate transporter 10 | SLC16A10 | 6q21-q22 | Q8TF71 | details | | Glutamate [NMDA] receptor subunit 3A | GRIN3A | 9q31.1 | Q8TCU5 | details | | Transient receptor potential cation channel subfamily M member 1 | TRPM1 | 15q13.3 | Q7Z4N2 | details | | Glycine receptor subunit alpha-1 | GLRA1 | 5q32 | P23415 | details | | Glycine receptor subunit alpha-2 | GLRA2 | | P23416 | details | | Glycine receptor subunit alpha-3 | GLRA3 | 4q33-q34 | O75311 | details | | Glycine receptor subunit beta | GLRB | 4q31.3 | P48167 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 1.125 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 0.345 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 2.725 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.316 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 0.604 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 1.037 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000162 |
|---|
| DrugBank ID | DB00172 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB000570 |
|---|
| KNApSAcK ID | C00001388 |
|---|
| Chemspider ID | 128566 |
|---|
| KEGG Compound ID | C00148 |
|---|
| BioCyc ID | PRO |
|---|
| BiGG ID | 34042 |
|---|
| Wikipedia Link | L-proline |
|---|
| METLIN ID | 29 |
|---|
| PubChem Compound | 145742 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17203 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
- Henzi V, Reichling DB, Helm SW, MacDermott AB: L-proline activates glutamate and glycine receptors in cultured rat dorsal horn neurons. Mol Pharmacol. 1992 Apr;41(4):793-801. [PubMed:1349155 ]
- Fichman Y, Gerdes SY, Kovacs H, Szabados L, Zilberstein A, Csonka LN: Evolution of proline biosynthesis: enzymology, bioinformatics, genetics, and transcriptional regulation. Biol Rev Camb Philos Soc. 2015 Nov;90(4):1065-99. doi: 10.1111/brv.12146. Epub 2014 Nov 4. [PubMed:25367752 ]
- Nadler JV, Wang A, Hakim A: Toxicity of L-proline toward rat hippocampal neurons. Brain Res. 1988 Jul 19;456(1):168-72. doi: 10.1016/0006-8993(88)90358-7. [PubMed:3409032 ]
- Mitsubuchi H, Nakamura K, Matsumoto S, Endo F: Inborn errors of proline metabolism. J Nutr. 2008 Oct;138(10):2016S-2020S. doi: 10.1093/jn/138.10.2016S. [PubMed:18806117 ]
|
|---|
