| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:23:15 UTC |
|---|
| Updated at | 2022-12-13 23:36:24 UTC |
|---|
| CannabisDB ID | CDB000059 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Histidine |
|---|
| Description | Histidine, or l-histidine, abbreviated His or H, an alpha-amino acid, is propanoic acid bearing an amino substituent at position 2 and a 1H-imidazol-4-yl group at position 3. Histidine is an essential amino acid that is not synthesized de novo in humans and must be obtained though ingestion of histidine. L-Histidine is a strong basic compound. It is an odorless white crystalline powder with a bitter taste. In humans, L-histidine is involved in methylhistidine metabolism. Histidine biosynthesis has been well studied in E. coli and involves eight gene products (His1, 2, 3, 4, 5, 6, 7, and 8, from a single gene) and occurs in ten steps. This is possible because a single gene product can catalyze more than one reaction (PMID:8852895 ). L-Histidine is found in high concentrations in beluga whales, caseins, and bearded seals and in lower concentration in prairie turnips, puddings, and black walnuts. L-Histidine has also been detected in climbing beans, sago palms, vanilla, pitanga (cherry) and allspices making histidine a potential biomarker for the consumption of these foods. Histidine is an amino acid found in cannabis plants (PMID: 6991645 ). It is a favouring ingredient and dietary supplement. Histidine was shown to have anti-hypertensive effects, decreasing blood pressure, by modulating the histamine H3 receptor (PMID: 19566844 ). Histidine is a precursor of histamine, an inflammatory product associated with allergies ( Ref:DOI ). Histidine can be post-translationally modified to 3-methylhistidine, which is excreted in urine and is a biomarker of skeletal muscle damage (PMID: 16079625 ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (S)-4-(2-Amino-2-carboxyethyl)imidazole | ChEBI | | (S)-alpha-Amino-1H-imidazole-4-propanoic acid | ChEBI | | (S)-alpha-Amino-1H-imidazole-4-propionic acid | ChEBI | | H | ChEBI | | His | ChEBI | | HISTIDINE | ChEBI | | L-(-)-Histidine | ChEBI | | L-Histidin | ChEBI | | (S)-a-Amino-1H-imidazole-4-propanoate | Generator | | (S)-a-Amino-1H-imidazole-4-propanoic acid | Generator | | (S)-alpha-Amino-1H-imidazole-4-propanoate | Generator | | (S)-Α-amino-1H-imidazole-4-propanoate | Generator | | (S)-Α-amino-1H-imidazole-4-propanoic acid | Generator | | (S)-a-Amino-1H-imidazole-4-propionate | Generator | | (S)-a-Amino-1H-imidazole-4-propionic acid | Generator | | (S)-alpha-Amino-1H-imidazole-4-propionate | Generator | | (S)-Α-amino-1H-imidazole-4-propionate | Generator | | (S)-Α-amino-1H-imidazole-4-propionic acid | Generator | | (S)-1H-Imidazole-4-alanine | HMDB | | (S)-2-Amino-3-(4-imidazolyl)propionsaeure | HMDB | | (S)-Histidine | HMDB | | (S)1H-Imidazole-4-alanine | HMDB | | 3-(1H-Imidazol-4-yl)-L-alanine | HMDB | | Amino-1H-imidazole-4-propanoate | HMDB | | Amino-1H-imidazole-4-propanoic acid | HMDB | | Amino-4-imidazoleproprionate | HMDB | | Amino-4-imidazoleproprionic acid | HMDB | | Glyoxaline-5-alanine | HMDB | | Histidine, L isomer | HMDB | | Histidine, L-isomer | HMDB | | L-Isomer histidine | HMDB | | L-Histidine | KEGG |
|
|---|
| Chemical Formula | C6H9N3O2 |
|---|
| Average Molecular Weight | 155.15 |
|---|
| Monoisotopic Molecular Weight | 155.0695 |
|---|
| IUPAC Name | (2S)-2-amino-3-(1H-imidazol-5-yl)propanoic acid |
|---|
| Traditional Name | L-histidine |
|---|
| CAS Registry Number | 30641-68-0 |
|---|
| SMILES | N[C@@H](CC1=CN=CN1)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H9N3O2/c7-5(6(10)11)1-4-2-8-3-9-4/h2-3,5H,1,7H2,(H,8,9)(H,10,11)/t5-/m0/s1 |
|---|
| InChI Key | HNDVDQJCIGZPNO-YFKPBYRVSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as histidine and derivatives. Histidine and derivatives are compounds containing cysteine or a derivative thereof resulting from reaction of cysteine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Histidine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Histidine or derivatives
- Alpha-amino acid
- L-alpha-amino acid
- Imidazolyl carboxylic acid derivative
- Aralkylamine
- Azole
- Imidazole
- Heteroaromatic compound
- Amino acid
- Carboxylic acid
- Azacycle
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Organic nitrogen compound
- Organooxygen compound
- Organonitrogen compound
- Primary amine
- Primary aliphatic amine
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Carbonyl group
- Organic oxygen compound
- Amine
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Indirect biological role: Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 287 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 45.6 mg/mL | Not Available | | logP | -3.32 | CHMELIK,J ET AL. (1991) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-001i-9100000000-ae0a0b81669663b3b4a5 | 2018-05-25 | View Spectrum | | GC-MS | Histidine, 3 TMS, GC-MS Spectrum | splash10-0udi-0940000000-43b9b035189af65e19cb | Spectrum | | GC-MS | Histidine, 3 TMS, GC-MS Spectrum | splash10-0udi-0910000000-fc3ecec9d4ff3aa8cb09 | Spectrum | | GC-MS | Histidine, non-derivatized, GC-MS Spectrum | splash10-0udi-0910000000-8284a673a35f7ac2241f | Spectrum | | GC-MS | Histidine, 3 TMS, GC-MS Spectrum | splash10-0fk9-8910000000-a3231b1a418b5798ecac | Spectrum | | GC-MS | Histidine, 3 TMS, GC-MS Spectrum | splash10-0udi-1920000000-227e8116769731104e82 | Spectrum | | GC-MS | Histidine, 4 TMS, GC-MS Spectrum | splash10-0ufu-2942100000-31605fd50ecc1f5be0ed | Spectrum | | GC-MS | Histidine, non-derivatized, GC-MS Spectrum | splash10-0udi-0940000000-43b9b035189af65e19cb | Spectrum | | GC-MS | Histidine, non-derivatized, GC-MS Spectrum | splash10-0udi-0910000000-fc3ecec9d4ff3aa8cb09 | Spectrum | | GC-MS | Histidine, non-derivatized, GC-MS Spectrum | splash10-0udi-0910000000-8284a673a35f7ac2241f | Spectrum | | GC-MS | Histidine, non-derivatized, GC-MS Spectrum | splash10-0002-2901100000-ae0f3865934743de764c | Spectrum | | GC-MS | Histidine, non-derivatized, GC-MS Spectrum | splash10-0fk9-8910000000-a3231b1a418b5798ecac | Spectrum | | GC-MS | Histidine, non-derivatized, GC-MS Spectrum | splash10-0udi-1920000000-227e8116769731104e82 | Spectrum | | GC-MS | Histidine, non-derivatized, GC-MS Spectrum | splash10-0ufu-2942100000-31605fd50ecc1f5be0ed | Spectrum | | GC-MS | Histidine, non-derivatized, GC-MS Spectrum | splash10-0ue9-0900000000-75e38a3f347210c301b0 | Spectrum | | GC-MS | Histidine, non-derivatized, GC-MS Spectrum | splash10-0udi-0910000000-c54bd92cc112eec7e509 | Spectrum | | Predicted GC-MS | Histidine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-001i-9500000000-1d29684ccb6383687865 | Spectrum | | Predicted GC-MS | Histidine, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-03di-6900000000-1dd400a16b8067f4daea | Spectrum | | Predicted GC-MS | Histidine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Histidine, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Histidine, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Histidine, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Histidine, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Histidine, TBDMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0a4i-0900000000-21530fac9975dc1edc7c | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-053r-9300000000-a946c375931adb366aae | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-053r-9000000000-61a3602d80bc0d54c579 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0a4i-0900000000-7d63b501889c2628f4f0 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-d4ccd4e44ca818b4cb17 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0a4i-0900000000-811aee5724fe0999c134 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0a4i-2900000000-052753eb699ae7cb4cb9 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0a4i-0900000000-3e145a05e6a71a0a2f56 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0a4i-0900000000-143a4b9d88183bc5abe7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-03di-0900000000-a8c213472da676a241f2 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-dfdf9562f155abb5fdeb | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0uk9-0879331100-8434b5b9e7e5700c2353 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-000i-0900000000-aaaaadcecd4cd7c94e1e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0udi-0900000000-90c4cbd09d7abc003013 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0udi-0900000000-1381708d18c24486773d | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0w29-0696321100-eb62fc608270d1151707 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-000i-0900000000-3450566ff38e6506a70f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-01q9-7900000000-124509a66476629ce10a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0udi-0900000000-efa6a21b1be16fda0802 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0udi-0900000000-137840e69da48c6114e4 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0udu-3900000000-06d50e90c797793a72d7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0006-9100000000-de7c9eb8068ef4f39f85 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-00l6-9000000000-ebf778679d4c6d6a6057 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-014i-9000000000-08d0188684283d1b2372 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0a4i-0900000000-46c2f608d27f0381a039 | 2012-08-31 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Histidine Metabolism | 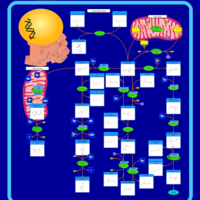 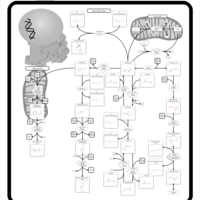 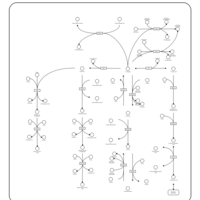 |  | | Ammonia Recycling |  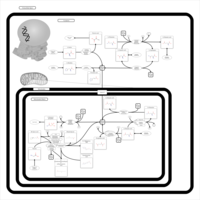 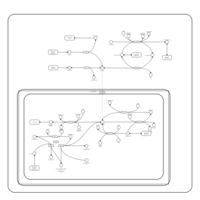 |  | | Transcription/Translation | Not Available | Not Available | | Beta-Alanine Metabolism | 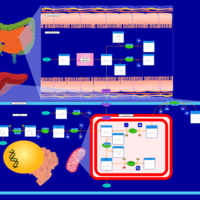 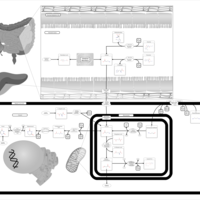 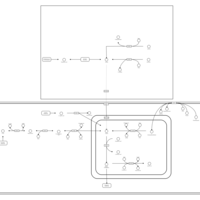 |  | | GABA-Transaminase Deficiency | 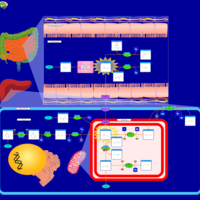 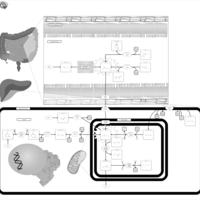 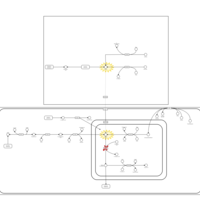 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
| Sodium-coupled neutral amino acid transporter 3 | SLC38A3 | 3p21.3 | Q99624 | details | | Monocarboxylate transporter 10 | SLC16A10 | 6q21-q22 | Q8TF71 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 0.673 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 0.875 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 1.0194 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.382 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 0.526 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 0.299 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000177 |
|---|
| DrugBank ID | DB00117 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB011856 |
|---|
| KNApSAcK ID | C00001363 |
|---|
| Chemspider ID | 6038 |
|---|
| KEGG Compound ID | C00135 |
|---|
| BioCyc ID | HIS |
|---|
| BiGG ID | 33985 |
|---|
| Wikipedia Link | L-histidine |
|---|
| METLIN ID | 21 |
|---|
| PubChem Compound | 6274 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15971 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
- Alifano P, Fani R, Lio P, Lazcano A, Bazzicalupo M, Carlomagno MS, Bruni CB: Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol Rev. 1996 Mar;60(1):44-69. [PubMed:8852895 ]
- Toba H, Nakamori A, Tanaka Y, Yukiya R, Tatsuoka K, Narutaki M, Tokitaka M, Hariu H, Kobara M, Nakata T: Oral L-histidine exerts antihypertensive effects via central histamine H3 receptors and decreases nitric oxide content in the rostral ventrolateral medulla in spontaneously hypertensive rats. Clin Exp Pharmacol Physiol. 2010 Jan;37(1):62-8. doi: 10.1111/j.1440-1681.2009.05227.x. [PubMed:19566844 ]
- Chinkes DL: Methods for measuring tissue protein breakdown rate in vivo. Curr Opin Clin Nutr Metab Care. 2005 Sep;8(5):534-7. doi: 10.1097/01.mco.0000170754.25372.37. [PubMed:16079625 ]
|
|---|


